

Background: Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of autoantibodies and immune complexes, leading to damage in multiple organs. Recently, studies have suggested that changes in the gut microbiota and associated metabolites may contribute to immunopathogenesis in SLE.
Objectives: This study aims to analyze the gut microbiota and related metabolites in stool and serum samples from Korean patients with SLE and explore their clinical significance.
Methods: The SLE patients included in this study were recruited from Seoul St. Mary’s Hospital and Yeouido St. Mary’s Hospital. The gut microbiota was analyzed through 16S rRNA sequencing of stool samples, and non-targeted metabolomic profiling was performed on stool and serum samples using gas chromatography (GC)-mass spectroscopy (MS) and liquid chromatography (LC)-MS. Clinical information, including demographics, medications, laboratory data, disease activity-related parameters, and dietary habits, was also collected from the SLE patients.
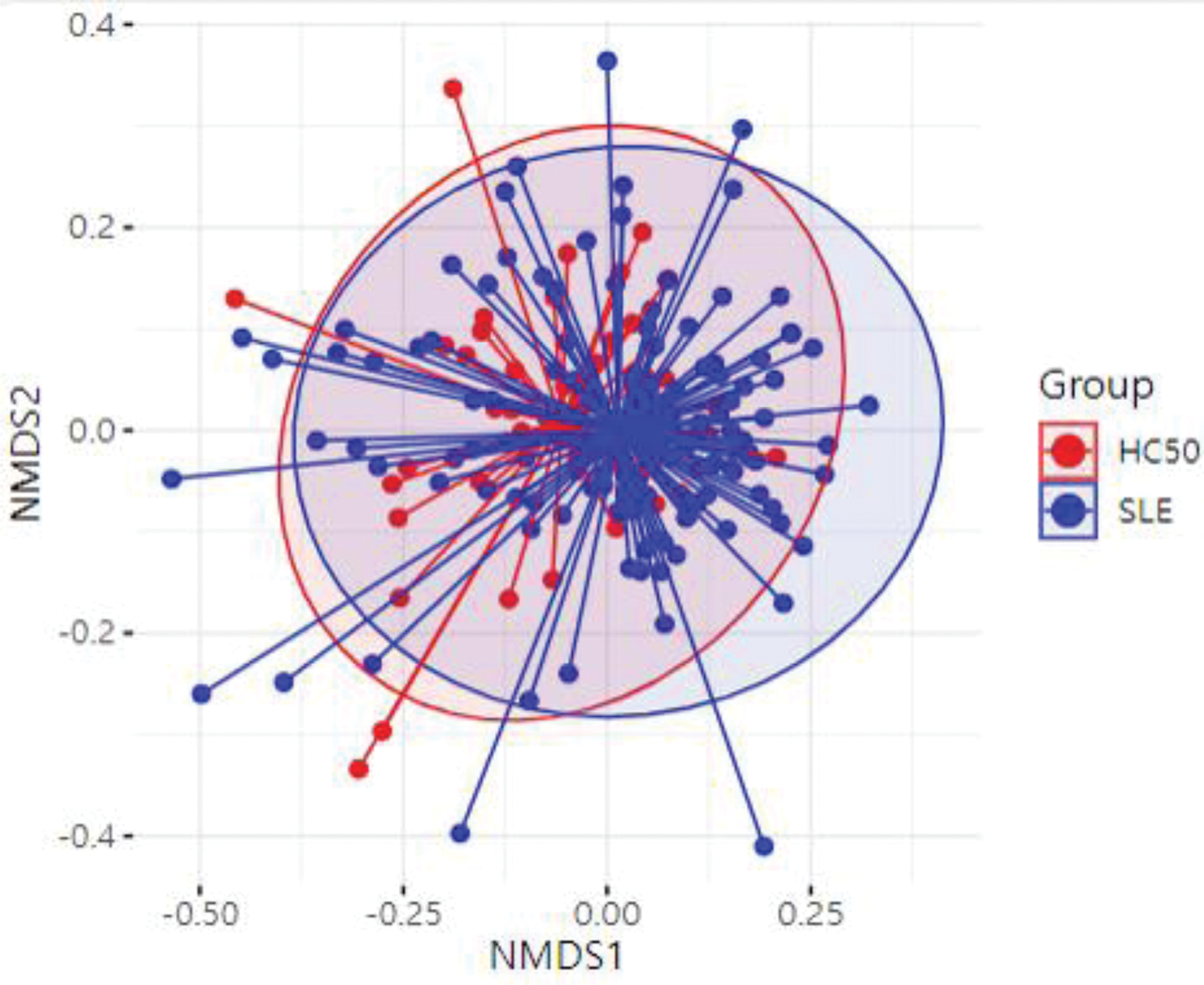
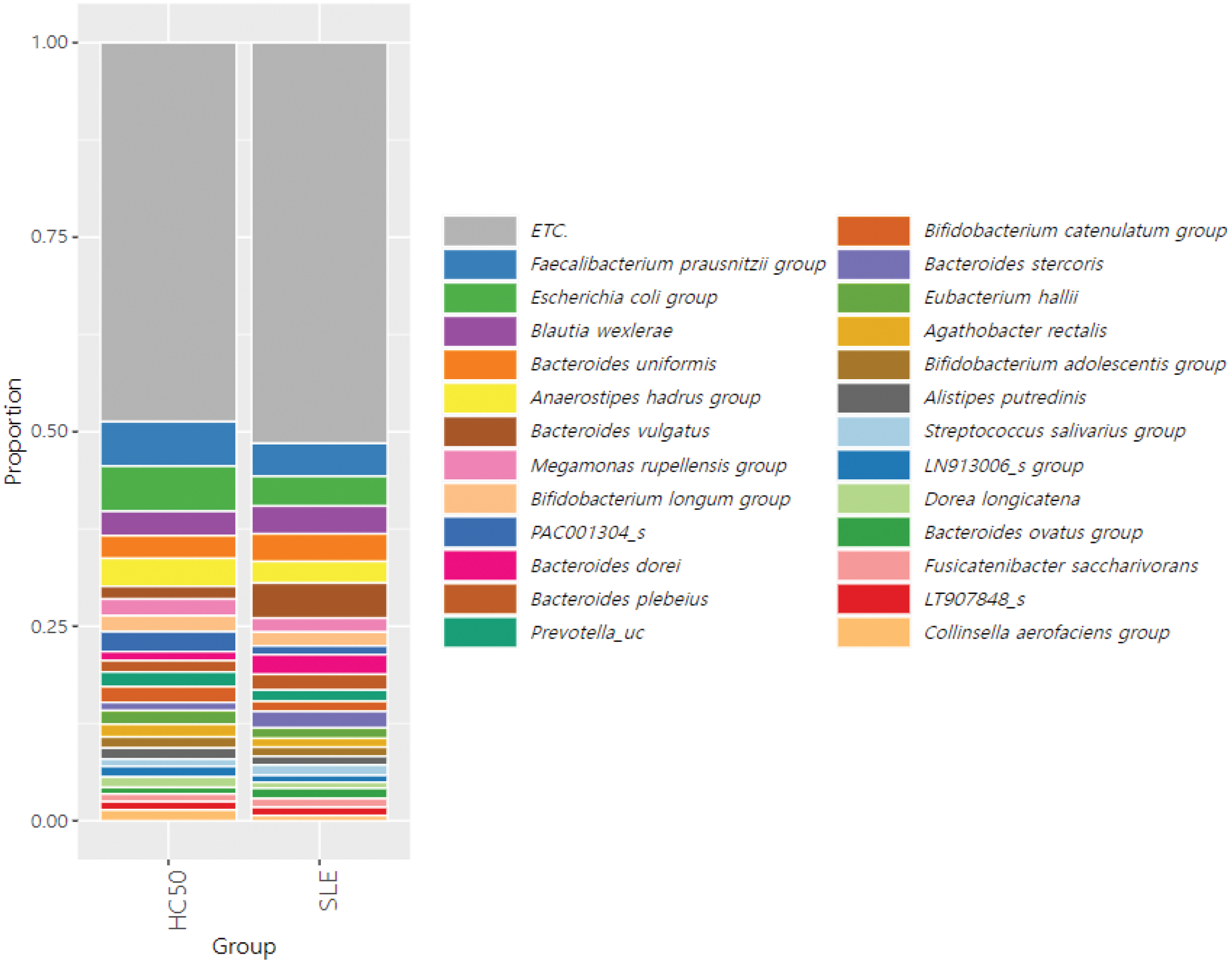
Results: 158 SLE patients and 50 healthy controls were recruited, with sample collection and clinical data acquisition completed. The median age of the SLE patients was 41 years, and 142 patients (89.9%) were female. Lupus nephritis was present in 65 patients (41.1%), and the median Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) score was 4. According to 16S rRNA sequencing analysis, SLE patients exhibited significant differences in diversity and composition of the gut microbiota compared to healthy controls. At the species level, Bacteroides vulgatus and Ruminococcus gnavus were enriched in the stool samples of SLE patients, whereas Faecalibacterium prausnitzii and Bifidobacterium bifidum were reduced. Differences in microbiota were further analyzed based on age, presence of lupus nephritis, disease activity, and medications. The distribution of gut microbiota showed no significant differences based on the presence of lupus nephritis, SLEDAI scores, or the use of gastrointestinal medications. However, when patients were stratified by age, distinct differences in gut microbiota composition were observed. Additionally, 76 metabolites in stool and 19 metabolites in serum were identified as significantly altered in SLE patients compared to healthy controls by GC-MS and LC-MS. Several metabolites, including linoleic acid, were decreased in SLE patients and showed correlations with the relative abundance of specific gut microbiota.
Conclusion: Korean SLE patients demonstrated distinct alterations in the gut microbiota and associated metabolites in stool and serum compared to healthy controls. Based on these findings, further research is anticipated to uncover novel mechanisms of lupus pathogenesis and identify potential therapeutic candidates.
REFERENCES: [1] Sliverman G J et al., The gut microbiome in systemic lupus erythematosus: lessons from rheumatic fever. Nat Rev Rheumatol. 2024 Mar;20(3):143-157.
Principal coordinates analysis of gut microbiome from SLE patients and healthy controls (HC50).
Taxonomic rank of gut microbiome at the species level from SLE patients and healthy controls (HC50).
Acknowledgements: This research was supported by the Bio&Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No. RS-2023-00224099).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (