

Background: Takayasu arteritis (TAK) is a rare and chronic systemic vasculitis that primarily targets large vessels, including the aorta and its main branches. The disease is characterized by an autoimmune-mediated inflammatory process that leads to vascular stenosis, occlusion, and aneurysm formation. Despite advances in understanding the immunopathogenesis of TAK, the molecular mechanisms underlying the involvement of key immune cell populations remain incompletely understood. Among these, CD4+ and CD8+ T cells are known to play pivotal roles in orchestrating inflammatory responses and tissue damage in TAK. However, the specific genes and pathways driving their contribution to TAK remain largely unexplored.
Objectives: This study aimed to investigate the transcriptional profiles of CD4+ and CD8+ T cells from TAK patients compared to healthy controls. By identifying Differentially Expressed Genes (DEGs) and enriched biological pathways, we sought to uncover the molecular mechanisms underlying T cell involvement in TAK pathogenesis and identify potential therapeutic targets.
Methods: Peripheral blood samples were collected from 8 TAK patients and 3 healthy controls. CD4+ and CD8+ T cells were isolated using flow cytometry and subjected to bulk RNA sequencing. Differential expression analysis was performed to identify significantly up-regulated and down-regulated genes. Gene Ontology (GO), KEGG, and Reactome pathway enrichment analyses were conducted to investigate the biological processes and signaling pathways associated with the DEGs. The results were visualized using volcano plots and enrichment maps to highlight key findings.
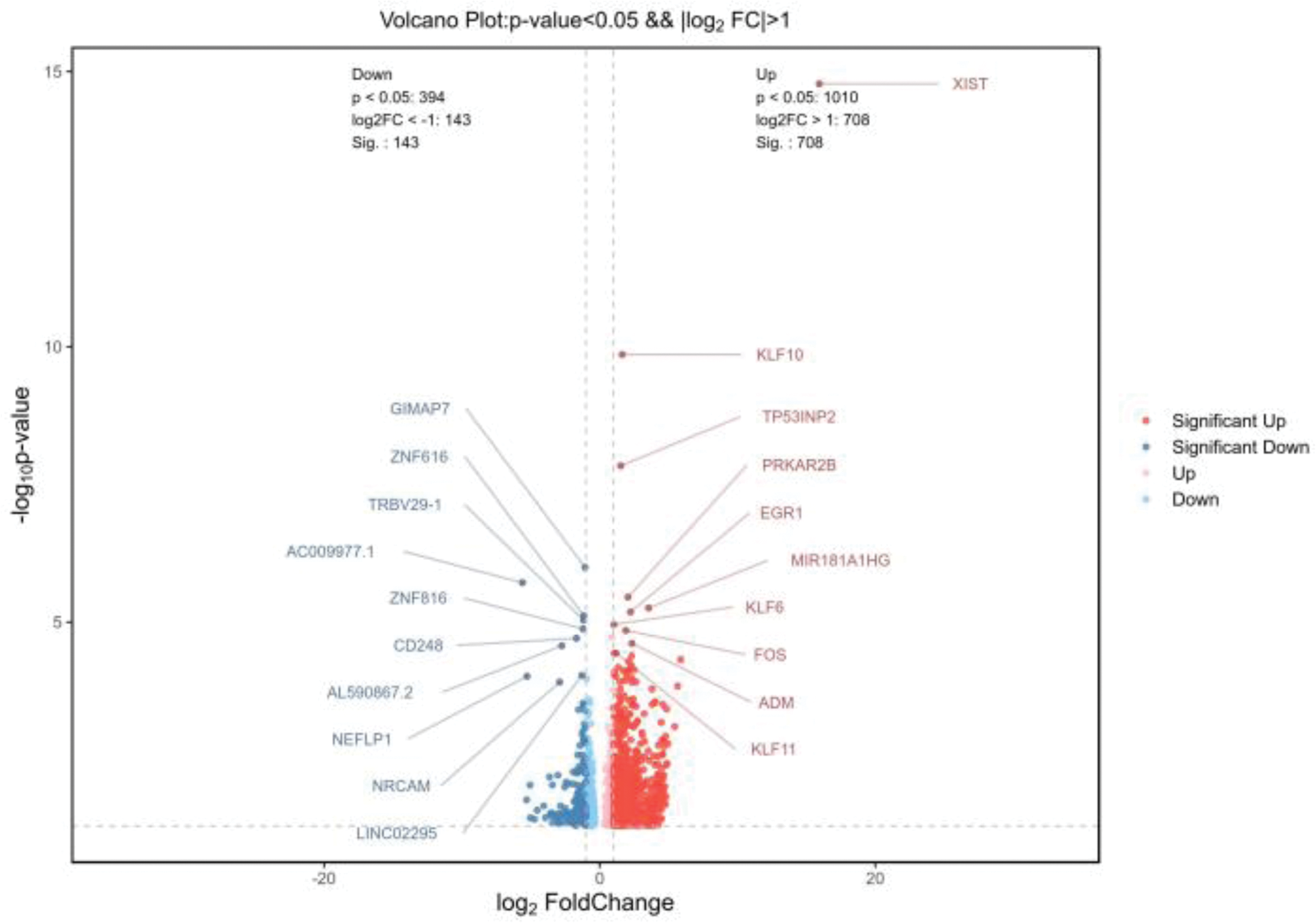
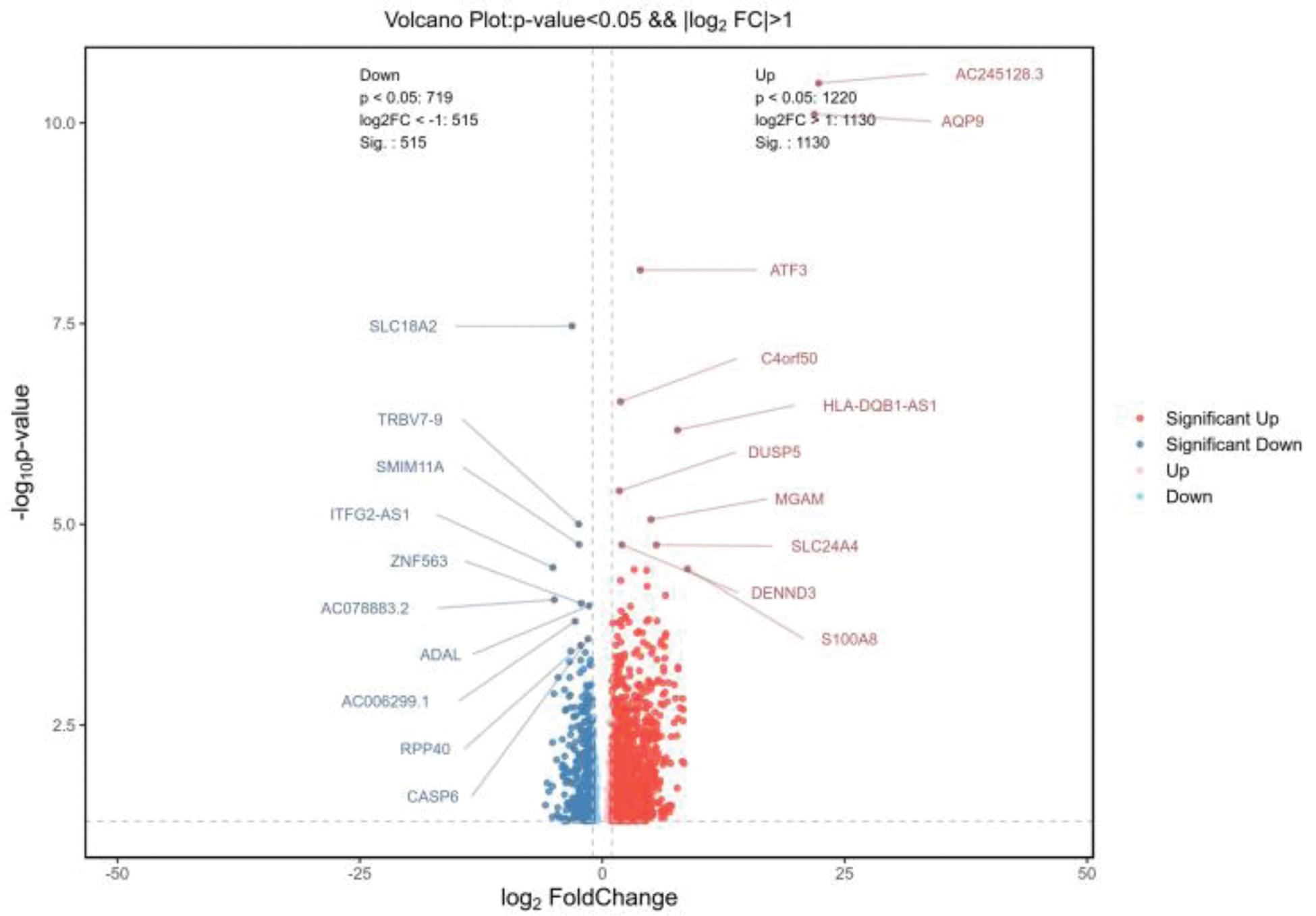
Results: 1. Bulk RNA sequencing revealed significant transcriptional differences in CD4+ and CD8+ T cells from TAK patients compared to healthy controls. In CD4+ T cells, 851 DEGs were identified, including up-regulated genes like XIST, KLF family members (KLF10, KLF11, KLF6), and EGR1, involved in immune regulation and inflammation, while down-regulated genes included GIMAP7 and CD248. In CD8+ T cells, 1645 DEGs were detected, with key up-regulated genes such as AQP9 and ATF3, and down-regulated genes linked to immune modulation and cytoskeletal dynamics (Figures 1 and 2). 2.Bulk RNA sequencing of CD4+ and CD8+T cells from TAK patients revealed significant enrichment in multiple GO pathways.CD4+ T cells in TAK were enriched in pathways related to inflammation, antibody responses, angiogenesis, and oxidative stress, with links to platelet-related structures, suggesting CD4+ T-cell–platelet interactions. Key functions included cytokine activity and chemokine receptor binding. CD8+ T cells showed enrichment in cytokines production, IL-1β regulation, and immune activation, with associations to membrane structures and roles in cell adhesion, signaling, and cytoskeletal regulation. These findings highlight the critical roles of CD4+ and CD8+ T cells in TAK pathogenesis. 3.KEGG pathway analysis of CD4+ T cells in TAK revealed key immune pathways like complement cascades, cytokines signaling, and TNF signaling, with platelet activation suggesting CD4+ T-cell–platelet interactions. CD8+ T cells showed similar patterns, including immune regulation and platelet activation. Shared enrichment in pathways like complement cascades and Rap1 signaling highlights the coordinated roles of both T-cell types in TAK pathogenesis. 4.Reactome pathway analysis of CD4+ T cells revealed significant enrichment in pathways like “platelet activation,” “degranulation,” “complement activation,” and “fibrin clot formation,” indicating a strong association with platelets in TAK. Additionally, enrichment in “chemokine receptor binding,” “GPCR ligand binding,” and “IL-4/IL-13 signaling” suggests roles in immune signaling and Th2-mediated inflammation.No particularly meaningful results were found in the Reactome pathways of CD8+ T cells.
Conclusion: Our study revealed significant transcriptional and pathway differences in CD4+ and CD8+ T cells from TAK patients, emphasizing their distinct roles in disease pathogenesis. CD4+ T cells demonstrated strong associations with immune and inflammatory responses, platelet activation, and Th2-mediated cytokine signaling, suggesting a key role in platelet-T cell interactions and vascular inflammation. In contrast, CD8+ T cells were primarily linked to cytokine production, immune activation, and cellular adhesion, with shared pathways highlighting coordinated functions between both T cell subsets. These findings provide novel insights into TAK immunopathology and potential therapeutic targets.
REFERENCES: NIL.
Volcano Plot Showing Differentially Expressed Genes in CD4+ T Cells of TAK Patients Compared to Healthy Controls.
Volcano Plot Showing Differentially Expressed Genes in CD8+ T Cells of TAK Patients Compared to Healthy Controls.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (