

Background: The role of neutrophils in the pathogenesis of Granulomatosis with polyangiitis (GPA) and Microscopic polyangiitis (MPA) is well established [1]. Whether the pattern of neutrophil activation differs between patients with newly diagnosed and relapsing disease is unclear.
Objectives: We sought to analyze the transcriptome of GPA/MPA patients with active disease or remission and investigate their molecular classification.
Methods: We conducted next-generation sequencing (101 bp single-end RNA-sequencing, NovaSeq6000-seq Illumina platform) in whole blood of patients with active GPA/MPA either at diagnosis (n=6), relapse (n=5) or remission (n=10; on Rituximab-maintenance therapy: n=4, off therapy: n=6) and age-/sex-matched healthy controls (n=12). None of patients with active disease received immunosuppressive therapy at the time of sampling. Sequences were aligned to the human genome (hg19; HISAT2 program); gene-expression quantification was performed by HTSeq and differential expression analysis was conducted using DESeq2, with FDR <0.05, log 2 FC<-1.5 and log 2 FC>1.5. Gene ontology analysis was performed using the EnrichR webtool.
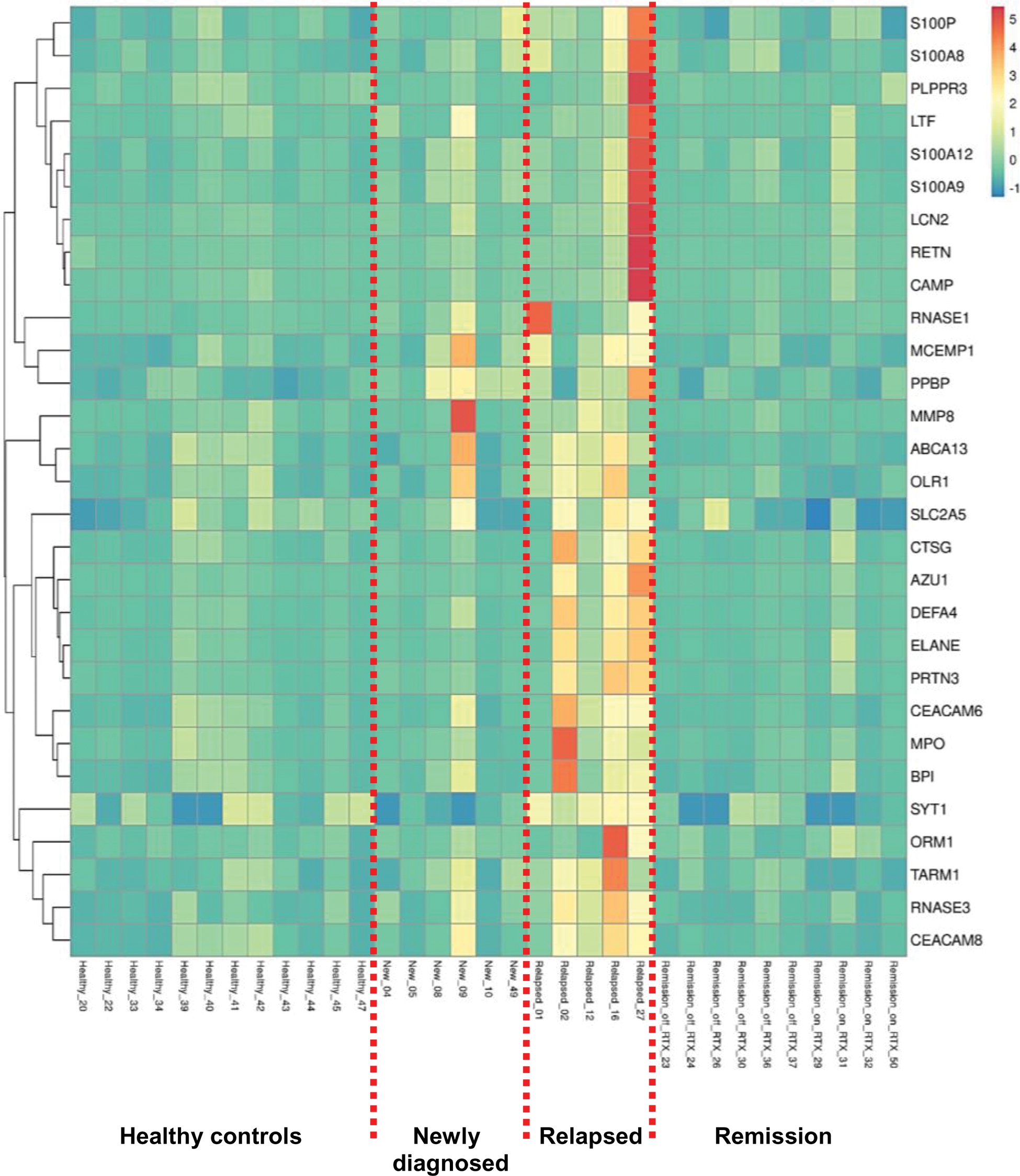
Results: Over-presentation analysis in relapsed compared to newly diagnosed patients with active GPA/MPA revealed upregulated molecular pathways relevant to neutrophil degranulation, neutrophil extracellular trap formation and antigen presentation. As a result, we discovered a unique gene signature comprised of 29 neutrophil-associated differentially expressed genes (Figure 1), which can distinguish relapsed patients from patients at diagnosis, remission and healthy controls.
Conclusion: There are clear differences in the neutrophil transcriptional reprogramming between newly diagnosed GPA/MPA patients compared to patients with relapsing disease. These findings could have implications in the better understanding of disease pathogenesis and treatment approaches.
Heatmap of neutrophil-associated DEGs in GPA/MPA patients with active disease (newly diagnosed or relapsed) or remission (on or off therapy) and in healthy controls. RTX; Rituximab, DEGs; differentially expressed genes.
REFERENCES: [1] Prendecki M, Gurung A, Pisacano N, Pusey CD. The role of neutrophils in ANCA-associated vasculitis. Immunol Lett. 2024;270:106933.
Acknowledgements: This work was supported in part by the Special Account for Research Grants, National and Kapodistrian University of Athens, Athens, Greece (DV #12085, 12086) and by the Greek Rheumatology Society and the Greek Association of Professional Rheumatologists (ERE-EPERE).
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (