

Background: Unstructured free-text data accounts for approximately 80% of electronic health records (EHR). Machine learning-based topic modeling (TM) techniques have been developed to extract insights and classify such unstructured data. While traditional natural language processing (NLP) models, mainly Latent Dirichlet Allocation (LDA), have been applied to topic modeling in the field of rheumatology, the adoption of advanced methods leveraging transformers and embeddings remains limited.
Objectives: To compare the performance of topic modeling techniques for identifying key themes in the EHR clinical notes of a rheumatology department.
Methods: One year clinical notes (January–December 2024) from a rheumatology department of a tertiary hospital were retrieved. These were de-identified and preprocessed by excluding those with <10 words, character count >3,000, or identified as potential duplicates. Embedded laboratory results were also removed using regular expressions. Two TM techniques were employed: LDA and BERTopic, a novel TM approach that combines transformer-based embeddings to capture semantic relationships among sentences with clustering algorithms. For LDA, preprocessing steps included the removal of stopwords, lemmatization, and the identification of bigrams using NLP techniques. For BERTopic, two embedding models were tested: “distiluse-base-multilingual-cased-v2” and “language-agnostic BERT sentence embedding (LaBSE)”, both well-suited for multilingual contexts as the notes were written in Catalan and Spanish. The minimum cluster size parameter of the HDBSCAN model was adjusted to identify the most effective model. Keyword selection was enhanced using Maximal Marginal Relevance (MMR) into the representation model for better topic interpretation. UMAP dimensionality reduction technique was used to visualise clinical notes and their assigned topics to explore semantic similarity. Coherence metrics (U-mass) were calculated to determine the optimal number of topics for LDA and overall model comparison. Finally, two expert rheumatologists reviewed and labeled the identified topics after analysing the associated keywords.
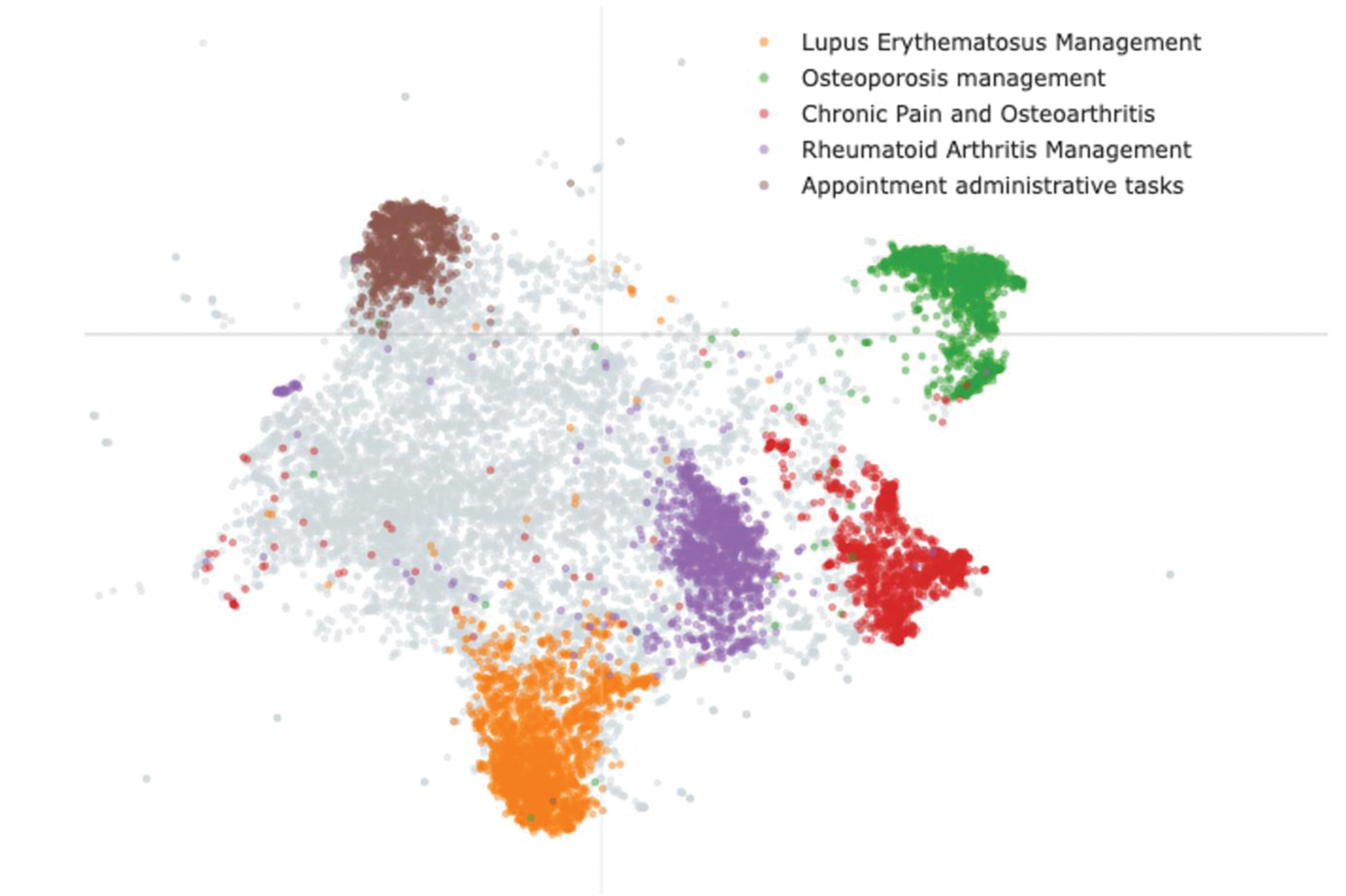
Results: A total of 26,789 clinical notes were retrieved, which were reduced to 21,418 following preprocessing. Among the tested LDA models, the model with 8 topics achieved the highest coherence score. From this model, two dominant topics emerged, collectively accounting for 62% (13,251/21,418) of the notes. Topic 1, labeled “Inflammatory Arthritis”, was characterised by keywords such as “pain,” “swelling,” “arthritis,” and classified 8,054 notes. Topic 2, labeled “Metabolic Bone Diseases and Osteoporosis”, included keywords such as “fractures,” “calcium,” and “DEXA,” and classified 5,197 notes. The BERTopic model using the “LaBSE” embeddings and a minimal cluster size of 50 retrieved the best results regarding coherence scores and clinical interpretability. Table 1 highlights the 10 most representative topics, with keywords translated to English. Figure 1 provides a visualization of topic distributions and the relationships among the 5 most prevalent topics, enabling a deeper understanding of topic similarity and note classification.
Conclusion: The BERTopic model demonstrated superior granularity and more effective labeling of topics compared to LDA. Key clinical topics identified in the electronic health record (EHR) rheumatology notes included Lupus , Osteoporosis , Rheumatoid Arthritis , and Chronic Pain Management . Notably, administrative appointment tasks constituted a significant portion of the notes. Clustering clinical notes by topic offers valuable insights, paving the way for future research into patient trajectories, treatment patterns, and clinical outcome.
Table 1. BERTopic model most prevalent 10 topics.
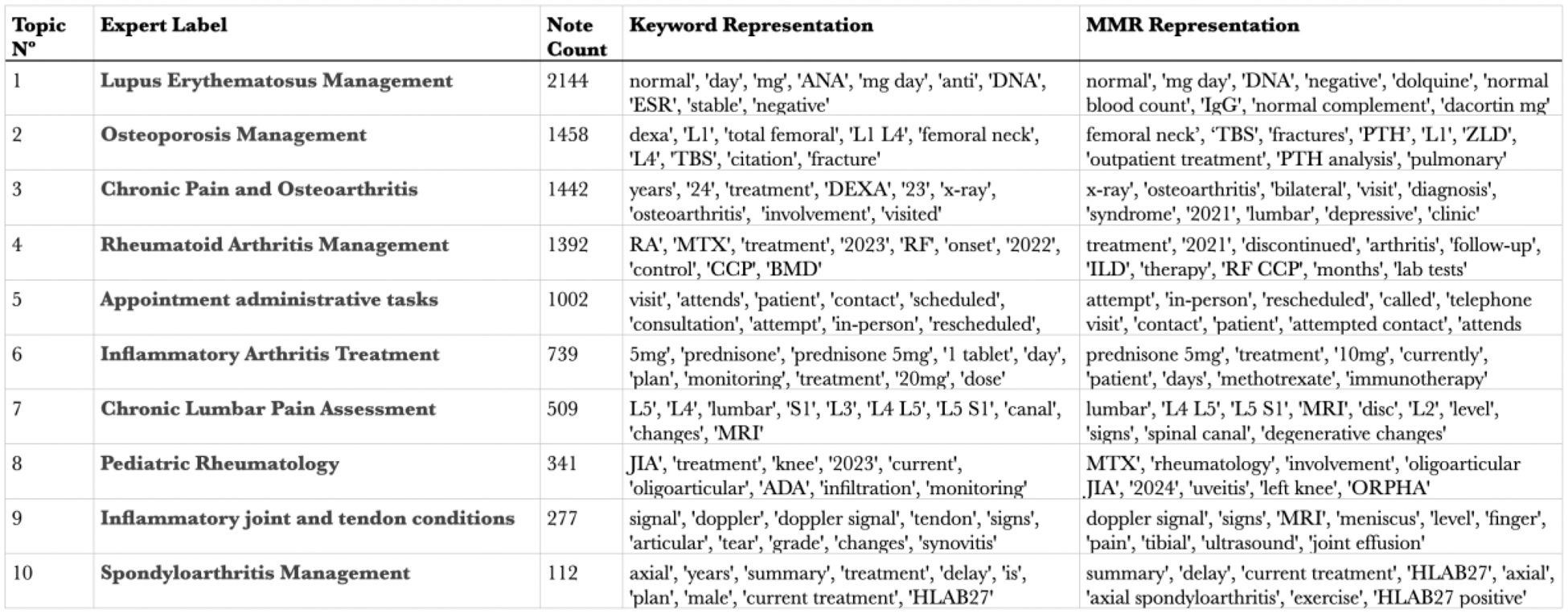
Visualization of document-topic relationships produced using BERTopic.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (