

Background: Large language models (LLMs) have demonstrated promise in clinical applications such as disease management, data extraction from electronic medical records, and decision support. However, ensuring accurate predictions remains challenging, especially for tasks that require precise clinical assessments, such as evaluating rheumatoid arthritis (RA) disease activity. Although cloud-based models (e.g., GPT-4) have shown potential, they pose data privacy and security concerns that limit their practical implementation. In contrast, open-source LLMs like Llama2 can be locally deployed to safeguard data, but they often lack specialized medical knowledge. Research on pre-training LLMs with medical literature and fine-tuning them for RA disease activity prediction has been limited, and direct comparisons with conventional statistical methods are lacking.
Objectives: The purpose of this study was to determine if Llama2 improves its predictive performance through additional medical pre-training (Meditron) and subsequent fine-tuning for RA disease activity at two years. The secondary purpose was to compare fine-tuned LLMs (Llama2 and Meditron) with a linear regression model to assess whether LLM-based predictions can surpass conventional approaches.
Methods: Clinical data from 11,865 Japanese patients with rheumatoid arthritis (RA) were obtained from the Institute of Rheumatology, Rheumatoid Arthritis (IORRA) cohort. This cohort originally included 15,004 patients; however, 3,139 patients were excluded due to insufficient follow-up data (less than two years). The remaining cases were divided into a training set (n = 9,500), a validation set (n = 102), and a test set (n = 2,263). Baseline characteristics, medication use, comorbidities, and four disease indices (DAS28-ESR, DAS28-CRP, CDAI, and J-HAQ) were utilized to predict disease activity and physical function at two years. Two large language models (LLMs) were evaluated: Llama2 (70B), an open-source model developed by Meta, and Meditron (70B), a pre-trained model developed by external researchers using medical datasets such as PubMed articles and clinical guidelines. Both LLMs underwent fine-tuning using the QLoRA method, which inserts low-rank trainable matrices into specific layers to preserve the integrity of the base model while minimizing computational resource requirements. A logistic regression model served as the reference for comparison. Model performance was primarily assessed using the area under the curve (AUC) from receiver operating characteristic (ROC) analyses, with statistical comparisons conducted using the DeLong test. Additional metrics, including precision, recall, F1 score, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), were also calculated.
Results: Pre-training alone significantly enhanced predictive accuracy. Meditron without fine-tuning (red line in Figure 1) provided accurate explanations of the RA indices and achieved higher AUC than Llama2 without fine-tuning (blue line in Figure 1) for DAS28-ESR >5.1, DAS28-ESR <2.6, DAS28-CRP <2.3, J-HAQ >2.5, and J-HAQ <0.5 (P <0.05). Fine-tuning resulted in significant improvements in AUC for Llama2 across all indices (P <0.05, green line vs. blue line in Figure 1) except CDAI >22, and for Meditron in DAS28-ESR <2.6, DAS28-CRP >4.1, DAS28-CRP <2.3 and CDAI ≤2.8 (P <0.05, black line vs. red line in Figure 1). Fine-tuned Llama2 (green line vs. yellow line in Figure 1) and Meditron (black line vs. yellow line in Figure 1) outperformed the linear regression model in terms of the baseline in DAS28-ESR <2.6, DAS28-CRP >4.1, DAS28-CRP <2.3, J-HAQ >2.5, and J-HAQ <0.5 (P <0.05). The LLMs, either Llama2 or Meditron outperformed the linear regression model across all metrics (sensitivity, specificity, PPV, NPV, precision, recall, and F1 scores) in DAS28-CRP >4.1, DAS28-CRP <2.3, J-HAQ score >2.5, and J-HAQ score <0.5 (Table 1). Furthermore, for the specificity and F1 score, Llama2 and Meditron consistently achieved higher values than the linear regression model across these indices (Table 1).
Conclusion: This study demonstrates that both pre-training on medical data and fine-tuning substantially improve Llama2 for predicting RA disease activity and physical function. Meditron’s pre-trained domain knowledge led to more accurate predictions prior to fine-tuning, while fine-tuned Llama2 performed comparably to fine-tuned Meditron in multiple categories. Both LLMs surpassed the linear regression model in key indices, underscoring their potential as secure, locally deployable decision support tools in clinical practice.
REFERENCES: NIL.
Comparison of predictive performance between the models across multiple indices.
Receiver operating characteristic (ROC) curves showing the classification performance of the five models predicting disease activity or physical function across three indices and J-HAQ scores in patients with rheumatoid arthritis: DAS28-ESR (A), DAS28-CRP (B), CDAI (C), and J-HAQ (D). The models include linear regression, Llama2 (with and without fine-tuning), and Meditron (with and without fine-tuning). Panels (A), (B), and (C) display ROC curves for high disease activity (left) and remission (right) for DAS28-ESR, DAS28-CRP, and CDAI, respectively. Panel (D) shows ROC curves for J-HAQ scores >2.5 (left) and <0.5 (right).
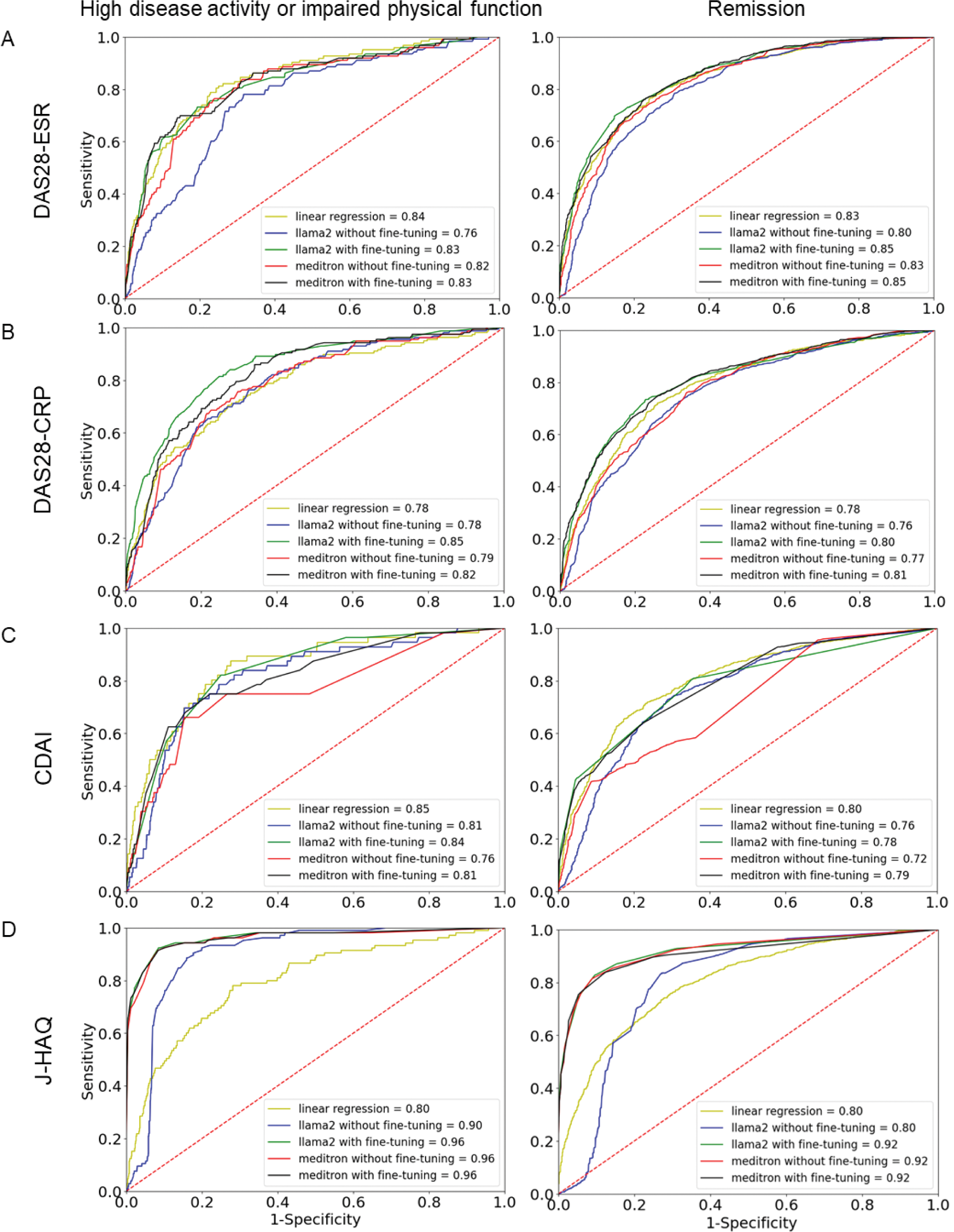
Table 1. Comparison of performance metrics between the linear regression model, Llama2, and Meditron across multiple disease indices
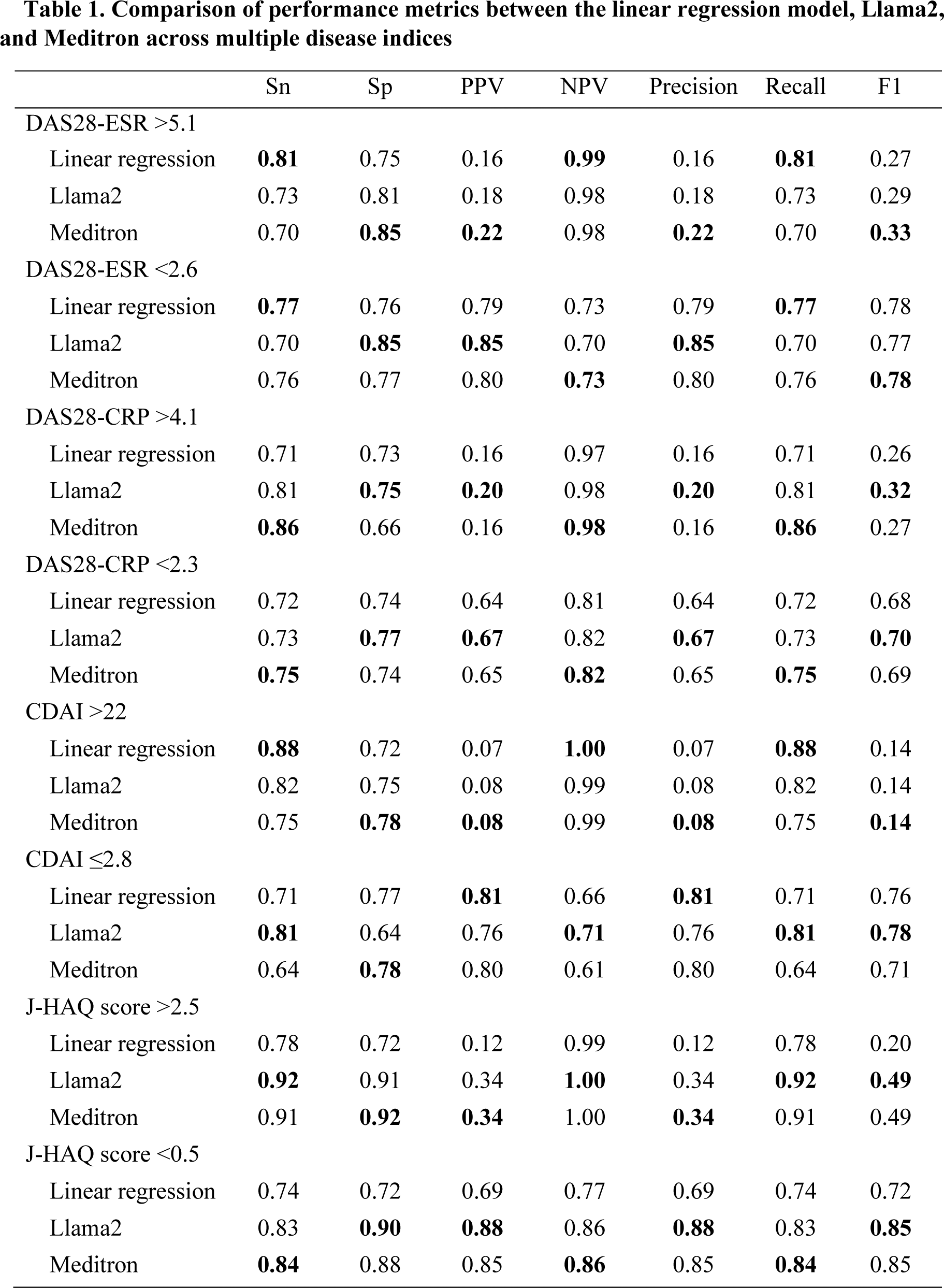
For each metric, the model with the best performance is highlighted in bold.
Sn, Sensitivity; SP, Specificity; PPV, positive predictive value; NPV, negative predictive value.
Acknowledgements: We thank all patients in the IORRA database and all members of the Institute of Rheumatology, Tokyo Women’s Medical University Hospital, for the successful management of the IORRA study cohort.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (