

Background: TREX1 is a negative regulator of the type I interferon response and TREX1 variants are widely considered to confer strong risk for non-monogenic lupus. The lack of validated instruments to capture peripheral type I interferon activity in large biobanks, however, limits the replication of these genetic associations. As such, there is a pressing need for a peripheral type I interferon signature measurement which can be used in large biobanks to adequately characterise immune endotypes and re-evaluate what is considered to be a canonical genetic association.
Objectives: We aimed to i) build a proteomic type I interferon signature, ii) define type I interferon-related autoimmunity, and iii) re-examine the association of rare TREX1 variants with non-monogenic lupus and type I interferon-related autoimmunity.
Methods: We used data from UK Biobank (UKB), a population-based study of 502k adult volunteers recruited in 2006-2010. We first built and validated the Markers of Interferon-α Response in Olink (MIRO) score defined from 12 interferon-stimulated gene products captured in UKB. Second, we assessed the association of MIRO with 20 autoimmune conditions to define type I interferon-related autoimmunity. Third, we leveraged this proteomic instrument to re-examine TREX1 lupus risk variants. Whole-exome sequencing was used to identify rare TREX1 variants reported as non-monogenic lupus risk factors or causing TREX1 -related monogenic disease in ClinVar, LOVD and prior publications. To further characterise 10 reported lupus risk variants, we assessed their functional impact using recombinant protein studies in Trex1 -/- mouse embryonic fibroblasts. We tested associations with Firth penalized logistic and linear regressions and replicated results in European ancestry participants using SKAT-O and deleteriousness predictions (LOFTEE). We finally performed a systematic review and meta-analysis of TREX1 -lupus genetic association studies using concepts “genetic variants”, “TREX1” and “lupus”.
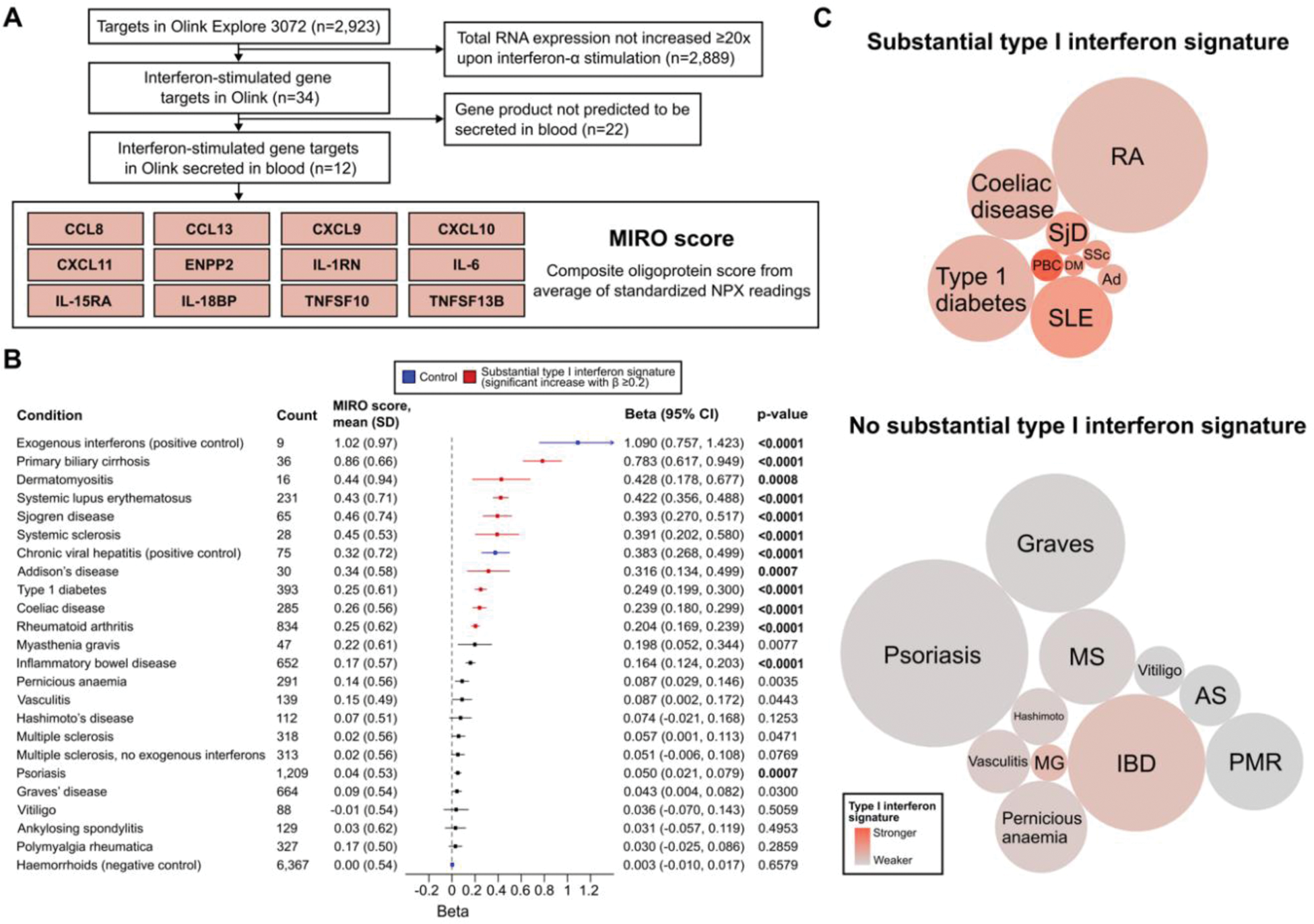
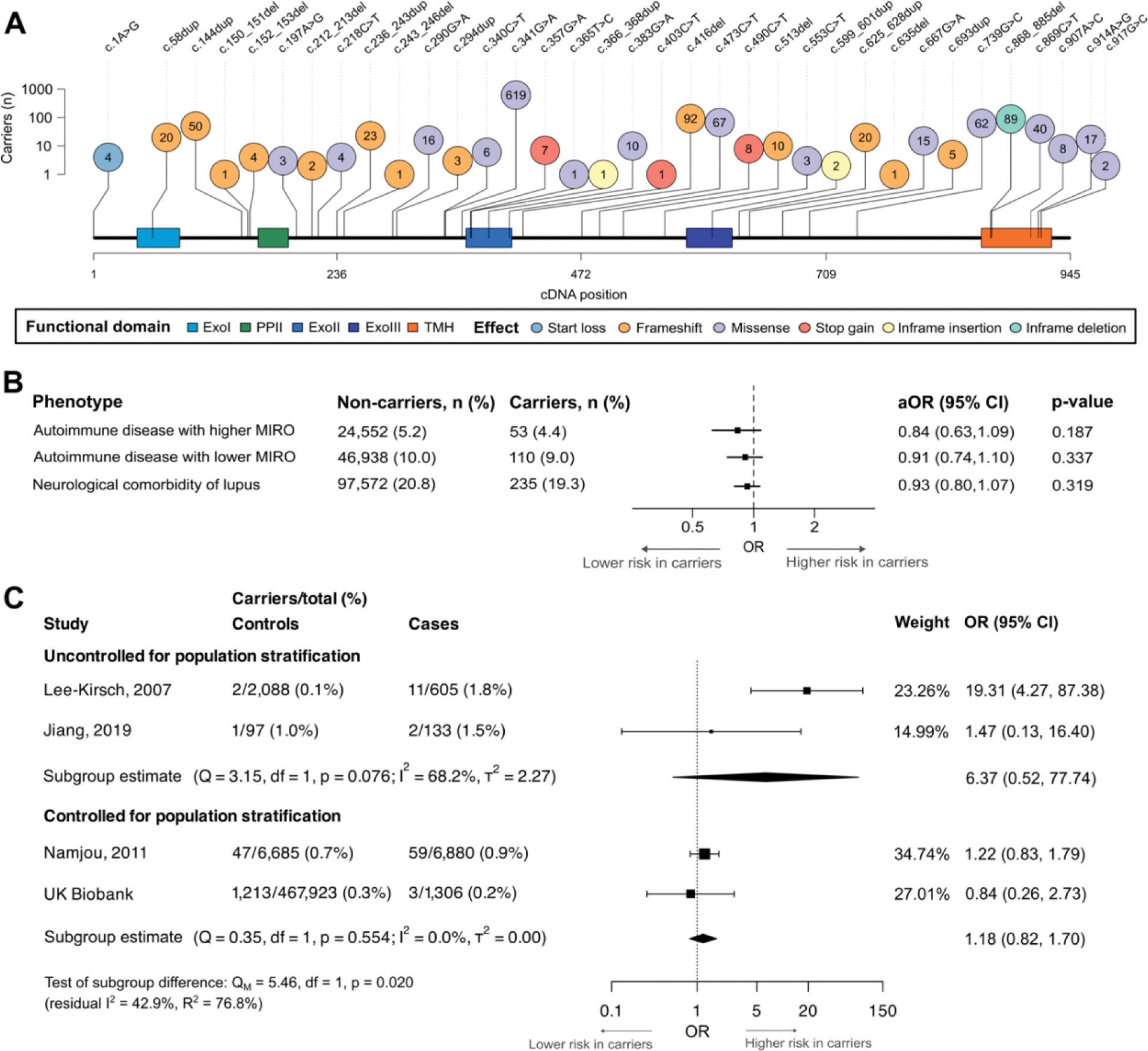
Results: A peripheral type I interferon signature (proxied by MIRO; Figure 1A) was observed in UKB participants on interferon therapy and with chronic viral hepatitis (positive controls) but not in those with haemorrhoids (negative control; Figure 1B). MIRO scores were markedly correlated with known interferon-stimulated genes (strongest correlation: galectin 9, r=0.692, p<0.0001). A substantial type I interferon signature was observed for nine conditions including lupus and Sjogren disease, which we defined as type I interferon-related autoimmunity (Figure 1C). These results were then used to comprehensively re-examine TREX1 lupus risk variants. Among 469,229 UKB participants, we identified 1,216 carriers (no homozygote) of any of 35 TREX1 variants (Figure 2A) and 1,306 lupus cases (69.5% from hospital admissions). Participants carrying any of the 10 previously reported TREX1 lupus risk variants did not have an increased risk of lupus (OR=0.178; 95% CI: 0.001, 1.207; p=0.090). In functional assays, only two of these 10 variants (R114H, P212Hfs) had any effect on TREX1 nuclease activity. Carriers of rare TREX1 variants did not have an increased risk of type I interferon-related autoimmunity, other autoimmune conditions or neurological comorbidities of lupus (Figure 2B), and did not have higher MIRO scores (β=0.01; 95% CI: -0.08, 0.10; p=0.829). These neutral results were reproduced with SKAT-O using very rare and high-confidence loss-of-function variants. In a meta-analysis of three published TREX1 -lupus studies and UKB data, we found no significant association of rare TREX1 variants with lupus (OR=2.16; 95% CI: 0.63, 7.34; p=0.219), with substantial heterogeneity (I 2 =76.6%) driven by two studies (including the first-in-topic candidate gene study) that did not account for population stratification (R 2 =76.8%; Figure 2C).
Conclusion: We built a proteomic type I interferon signature (the MIRO score) to identify type I interferon-related autoimmune diseases in UKB. Leveraging this proteomic signature, we found no significant association between TREX1 variants, non-monogenic lupus or type I interferon-related autoimmunity in UKB. Taken together, these results do not support the reputed risk of lupus conferred by rare TREX1 variants which has become embedded in narrative reviews. At a practical level, we suggest there is no merit in sequencing TREX1 in patients with lupus, where monogenic disease is not suspected.
Derivation and validation of a proteomic interferon score and identification of interferonopathic autoimmune diseases in UK Biobank. Panel A : Flowchart of Olink Explore 3072 targets in MIRO score. Panel B : Peripheral type I interferon activation using MIRO in linear regressions adjusted for age and sex. P-values with bold characters indicate statistical significance (p<0.0021 or 0.05/24 tests). Scores available for 44,843 UKB participants. Panel C : Type I interferon-related autoimmune diseases (significant increase with β≥0.2; colour: β, diameter: sample size). Abbreviations : Ad, Addison’s disease; AS, ankylosing spondylitis; DM, dermatomyositis; IBD, inflammatory bowel disease; MG, myasthenia gravis; MS, multiple sclerosis; NPX, normalized protein expression; PBC, primary biliary cirrhosis; PMR, polymyalgia rheumatica; RA, rheumatoid arthritis; SjD, Sjogren disease; SLE, systemic lupus erythematosus; SSc, systemic sclerosis.
Association of rare TREX1 variants with autoimmunity in UK Biobank and systematic review. Panel A : Lollipop plot of carrier counts in UK Biobank for rare TREX1 variants. Panel B : Clinical associations of rare TREX1 variants in UK Biobank. Panel C : Forest plot of previously reported TREX1 -lupus genetic association studies meta-analysed with UK Biobank results (random effects).
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (