

Background: Skin fibrosis is a hallmark of systemic sclerosis (SSc). It is established that a subset of SSc patients experience spontaneous regression of skin fibrosis. The molecular mechanisms underlying this resolution remain unclear. CD4 + T cell abnormalities have been linked to the vascular lesions and fibrosis in SSc through the altered release of pro-fibrotic and pro-inflammatory cytokines.
Objectives: This study investigates the alterations in CD4 + T cells in patients with regression of skin fibrosis versus those who progress.
Methods: Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation from 10 SSc patients. Regressors (N = 5) were defined by a 5-point decrease in the modified Rodnan Skin Score (mRSS), whereas progressors (N = 5) demonstrated a 5-point increase in mRSS at 1-year follow-up. Single-cell libraries were prepared using a droplet-based system (10X Genomics) and sequenced with the Illumina NovaSeq 6000. Raw reads were aligned to the reference human genome using Cellranger 7.0. Transcriptomic data were analyzed using the Seurat package (5.1.0). 36,574 cells were annotated to the major PBMC cell types (monocytes, CD4 + T cells, CD8 + T cells, natural killer cells, B cells, other T cells, and dendritic cells) and subsequently subset to include only CD4 + T cells (N = 6,248 cells). Differential gene expression analysis (DGEA) was performed using MAST with patients as a covariate to account for patient-driven effects. DGEA was performed on all genes expressed in >5% of cells and filtered for genes with an FDR < 0.3 and Log2FC |0.2|, followed by Gene-Set Enrichment Analysis (GSEA). For each gene set, a module score was generated using the AddModuleScore function by aggregating the average expression of each gene in the gene set. Pseudotime trajectory analysis was performed using Monocle3.
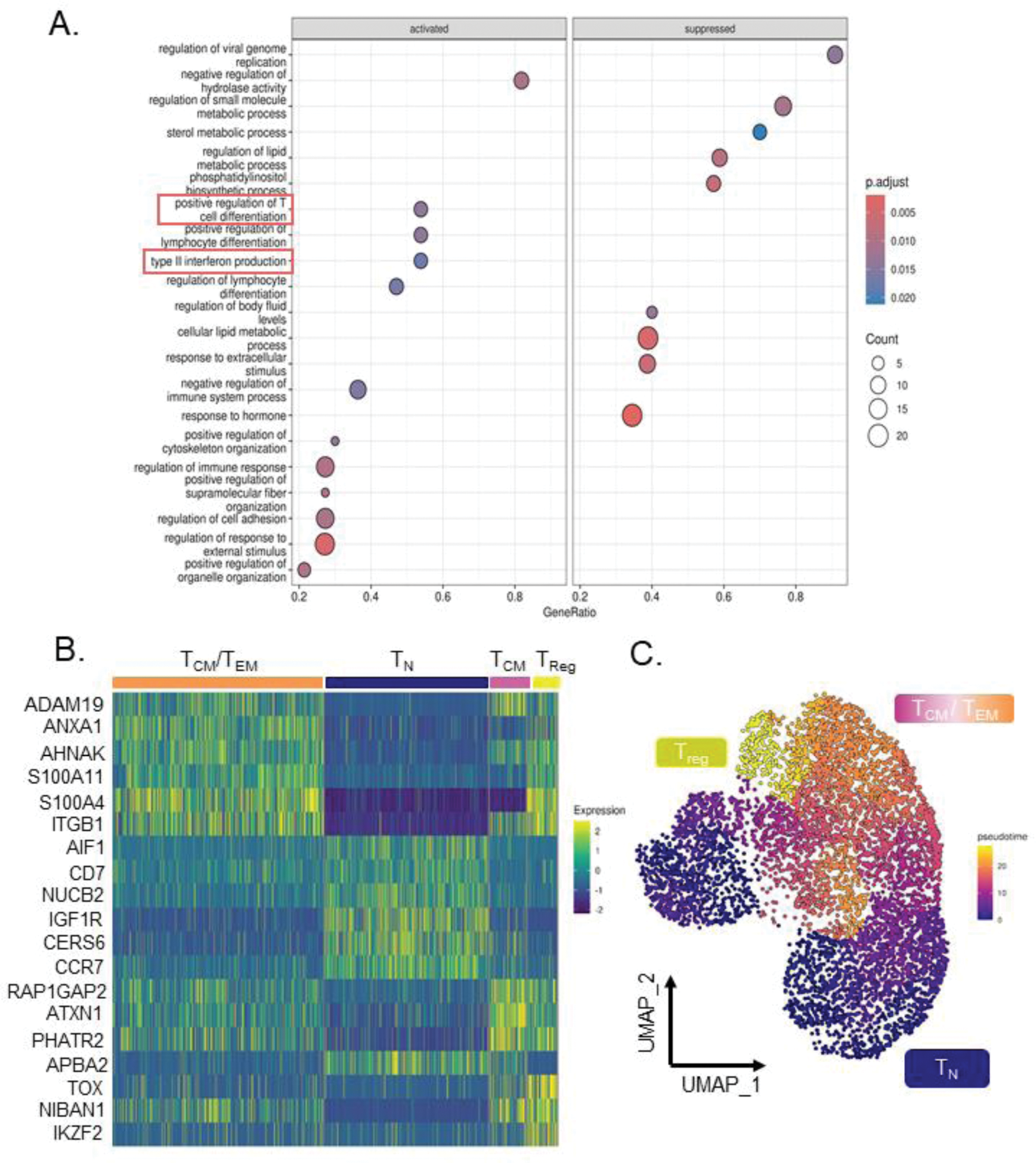
Results: DGEA was performed between regressors and progressors, yielding 410 of upregulated and 412 of downregulated genes in the regressors. GSEA using Gene Ontology (GO) Biological Processes revealed “Positive Regulation of T cell Differentiation” and “Type II Interferon (IFN-γ) Production” were activated in the regressors and showed increased gene module scores (Figure 1A). The “Positive Regulation of T cell Differentiation” gene set contained transcription factors GATA3 (Log2FC = 0.35), RUNX3 (Log2FC = 0.35), and FOXO3 (Log2FC = 0.33). GATA3 is the master regulator of naïve (T N ) to Th2 cell differentiation. RUNX3 inhibits production of IL-4, serving as a positive regulator of GATA3 expression. FOXO3 is a key regulator of regulatory T cell (T reg ) differentiation via activation of FOXP3. Reclustering of the subset CD4 + T cells generated 7 subclusters. Clusters 0, 1, 4 exhibited an enrichment for the IFN-γ production and Positive Regulation of T cell Differentiation module scores relative to the other clusters. Additionally, the gene module scores for these clusters were also increased in the regressors. GATA3, RUNX3, and FOXO3 also demonstrated increased expression in clusters 0, 1 and 4. The top 20 markers for each cluster were used for manual annotation of the subclusters (Figure 1B). Established T cell activation markers such as CD69 and IL2RA were used to assess the activation state of the clusters. Together, this resulted in an annotation of 4 subsets of CD4 + T cells: T N , central memory (T CM ), (memory) T regs , and T CM /effector memory (T EM ) cells. The subsets were further confirmed using pseudotime trajectory analysis, which shows the expected differentiation lineage from T N to T regs (Figure 1C). Clusters annotated as T CM /T EM (clusters 0 and 1) also displayed higher expression of the early activation marker CD69 . CD69 was among the top differentially expressed genes increased in the regressors, indicating increased T cell activation. Additionally, the module score for IFN-γ also localized to the T CM /T EM cluster. IFN-γ is secreted by activated T cells, particularly Th1 cells, and counteracts pro-fibrotic Th2 cytokines.
Conclusion: Our analysis suggests that SSc patients with skin fibrosis regression have an increased IFN-γ signature and T cell activation. This indicates that activated, pro-inflammatory CD4 + T cells in SSc patients may play a role in the regression of skin fibrosis.
(A ) Gene Set Enrichment Analysis using GO Biological processes, showing “Type II Interferon Production” and “Positive Regulation of T Cell Differentiation” as significantly increased in regressors compared to progressors. (B ) Heatmap showing the top 5 markers for each classification of the subclusters. (C ) Pseudotime analysis confirms the trajectory of T cell differentiation is in line with the subcluster annotation. Abbreviations: T N : Naïve T cells, T CM : Central Memory T cells, T EM : Effector Memory T cells, T reg : Regulatory T cells.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Astrid Hofman: None declared, Pietro Bearzi: None declared, Elena Pachera: None declared, Lumeng Li: None declared, Laura Much: None declared, Kristina Buerki: None declared, Anna-Maria Hoffmann-Vold Boehringer Ingelheim, Janssen, Medscape, Merck Sharp & Dohme, Novartis and Roche, AbbVie, ARXX, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Janssen, Medscape, Merck Sharp & Dohme, Pliant Therapeutics, Roche and Werfen, Boehringer Ingelheim, Janssen, Mike O. Becker GSK, Amgen, Novartis, Vifor, Amgen, Vifor, Foundation for research in Rheumatology (FOREUM), EMDO Foundation, Novartis foundation for medical-biological research, Innovative Medicines Initiative (IMI), Oliver Distler Bayer, Boehringer Ingelheim, Janssen, Medscape, 4P-Pharma, Abbvie, Acceleron, Alcimed, Altavant Siences, Amgen, AnaMar, Arxx, AstraZeneca, Baecon, Blade, Bayer, Boehringer Ingelheim, Corbus, CSL Behring, Galapagos, Glenmark, Horizon, Inventiva, Kymera, Lupin, Miltenyi Biotec, Mitsubishi Tanabe, MSD, Novartis, Prometheus, Redxpharna, Roivant, Sanofi and Topadur, BI, Kymera, Mitsubishi Tanabe, UCB.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (