

Background: Systemic lupus erythematosus (SLE) is an autoimmune disease with heterogeneous manifestations and a complex multifactorial etiology that stems from malfunctions in immune cells of both the innate and adaptive immune systems. Data regarding the dysregulation of immune system functions and pathways in patients with SLE who are not treated with immunosuppressive therapy is scarce.
Objectives: To describe immune signatures of SLE patients who have not received any treatment (newly diagnosed), and those currently treated with hydroxychloroquine (HCQ) only (established disease) compared to healthy controls, at baseline and over time, utilizing multi-omic single-cell RNA-sequencing. Additionally, to characterize immune signatures that are associated with different SLE manifestations.
Methods: SLE patients (≥18 years), classified according to SLICC 2012 and/or ACR/EULAR 2019 classification criteria, naïve or treated with HCQ alone were included in the study. Demographic and clinical data were collected from the patients and their medical records. Blood samples were obtained at baseline and after 3 and 6 months. Healthy control data were provided from the Immunai database. Peripheral blood monocytes (PBMCs) were isolated from all blood samples and analyzed at Immunai’s laboratory by single-cell RNA sequencing and cellular indexing of transcriptomes and epitopes (CITE) sequencing, annotation of cell subsets, and expression analysis.
Results: Eleven SLE patients (39 PBMC samples) and 12 healthy controls were included in the analysis. Of the SLE patients, 10 were female, with a mean age of 43.18±9.96 years. The mean SLEDAI-2K score at baseline was 4.55±4.36. Three patients had an SLE damage index of ≥1. Nine patients were treated with HCQ from baseline, 4 of whom had a past history of lupus nephritis. All SLE patients had baseline and 3-month samples, and five also had 6-month samples. We recovered a total of 156,168 high-quality cells, comprising 32 subtypes (Figure 1a). Upon examination of pathway expression scores, as expected, SLE patients had a higher interferon (IFN) response gene signature expression in all immune cell groups compared to controls (Figure 1b). In patients treated with HCQ, the IFN signature score decreased over time (Figure 1c). Interestingly, oxidative phosphorylation (oxphos) pathways from three different databases were the most enriched pathways in SLE patients compared to controls (Figure 2a). As with the IFN pathway, oxphos pathways decreased during HCQ treatment, while TNF pathways increased over time. We next analyzed differences in patients based on three clinical parameters - elevated anti-dsDNA antibodies, musculoskeletal manifestations, and history of nephritis. In SLE patients with elevated anti-dsDNA antibodies (n=7 at baseline, n=6 at 3 months) or musculoskeletal manifestations (n=5 at both time points), we found increased abundance of naïve CD4 and CD8 T-cells, while gamma-delta T-cells were less abundant (Figure 2b-c). In patients with a history of lupus nephritis (n=4), class-switched memory B cells, naïve CD4 and CD8 cells, and T-regs were more abundant, and gamma-delta T-cells, non-classical monocytes, and pDCs were less abundant (Figure 2d). At a transcriptional level, patients with elevated anti-dsDNA antibodies displayed increased protein synthesis activity in multiple B cell populations. In patients with active musculoskeletal manifestations, B-cell protein synthesis was decreased; CD4 T-cell subsets had upregulated TNFα, IFNγ, and IL-6 pathways; gamma delta T-cells showed upregulation of IL-10, IFNβ, and IFNγ pathways; and NK cell cytotoxic pathways were increased. In patients with a history of nephritis, a high inflammatory immune signature was observed with IFNα in almost all cell types. IFNβ and IFNγ pathways were also upregulated in many cell types.
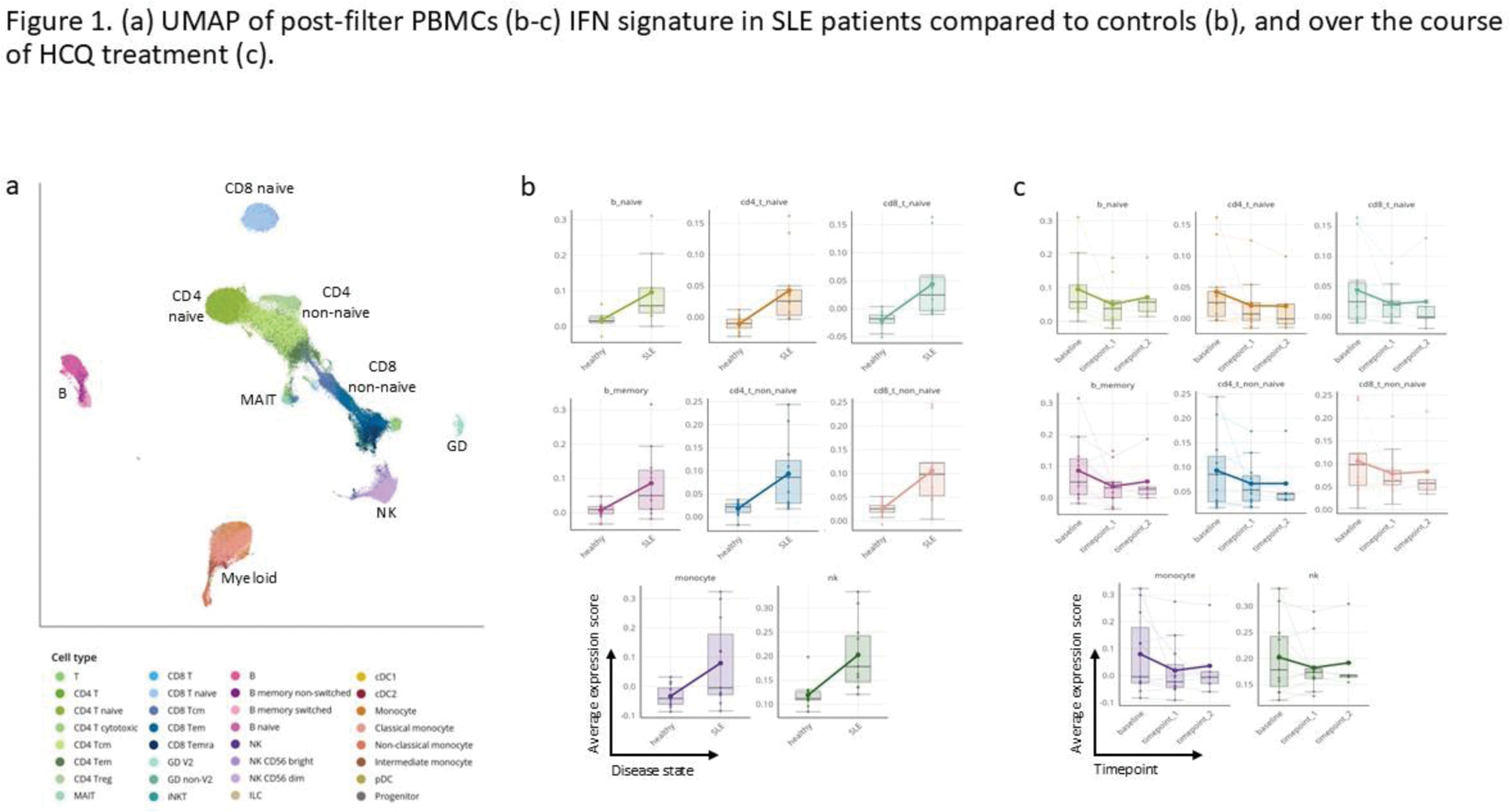
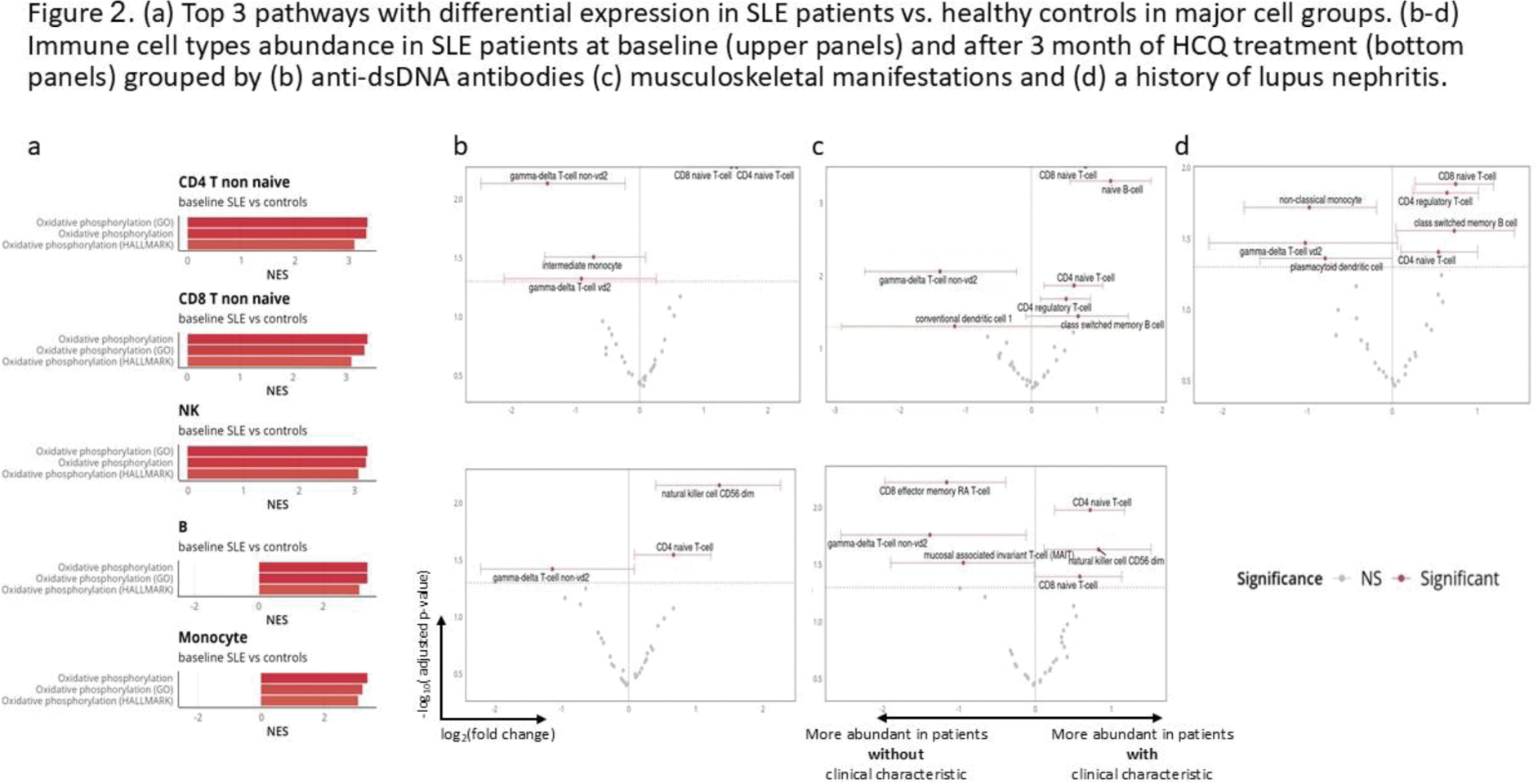
Conclusion: SLE patients have elevated expression of IFN and oxphos pathways across cell types as compared to healthy controls. Expression of these pathways decreases with HCQ treatment. Serologically or clinically active patients, as well as patients with a history of nephritis, show a similar immune cell abundance pattern which is different compared to patients with inactive disease.
REFERENCES: NIL.
Acknowledgements: The study was supported and performed in collaboration with Immunai Inc.
Disclosure of Interests: Tali Eviatar: None declared, Natan Stein Immunai Inc., Alexandra Tsitsiklis Immunai Inc., Gillian Mbambo Immunai Inc., Cailin Joyce Immunai Inc., Jonathan Wollman: None declared, Victoria Furer: None declared, Ari Polachek: None declared, Ori Elkayam: None declared, Daphna Paran: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (