

Background: Type I interferons play an important role in the pathogenesis of systemic lupus erythematosus (SLE), and anifrolumab (ANI), a human monoclonal antibody targeting type I interferon receptor subunit 1, has recently been approved for the treatment of SLE. Sub-analyses from the phase III TULIP trials (NCT02446912, NCT02446899) demonstrated that the efficacy of ANI was associated with reductions in interferon-stimulated gene (ISG) expression 12 weeks after treatment. SLE is characterized by various changes in the immune system, and in the B cell compartment, patients with SLE exhibit alterations in immunoglobulin heavy chain gene usage changes and clonal expansion. However, the specific cellular targets of ANI and its impact on the B cell receptor (BCR) repertoire remain poorly understood.
Objectives: The aim of this study was to evaluate the impact of type I interferon inhibition therapy on immune cells and the BCR repertoire using single-cell sequencing of peripheral blood.
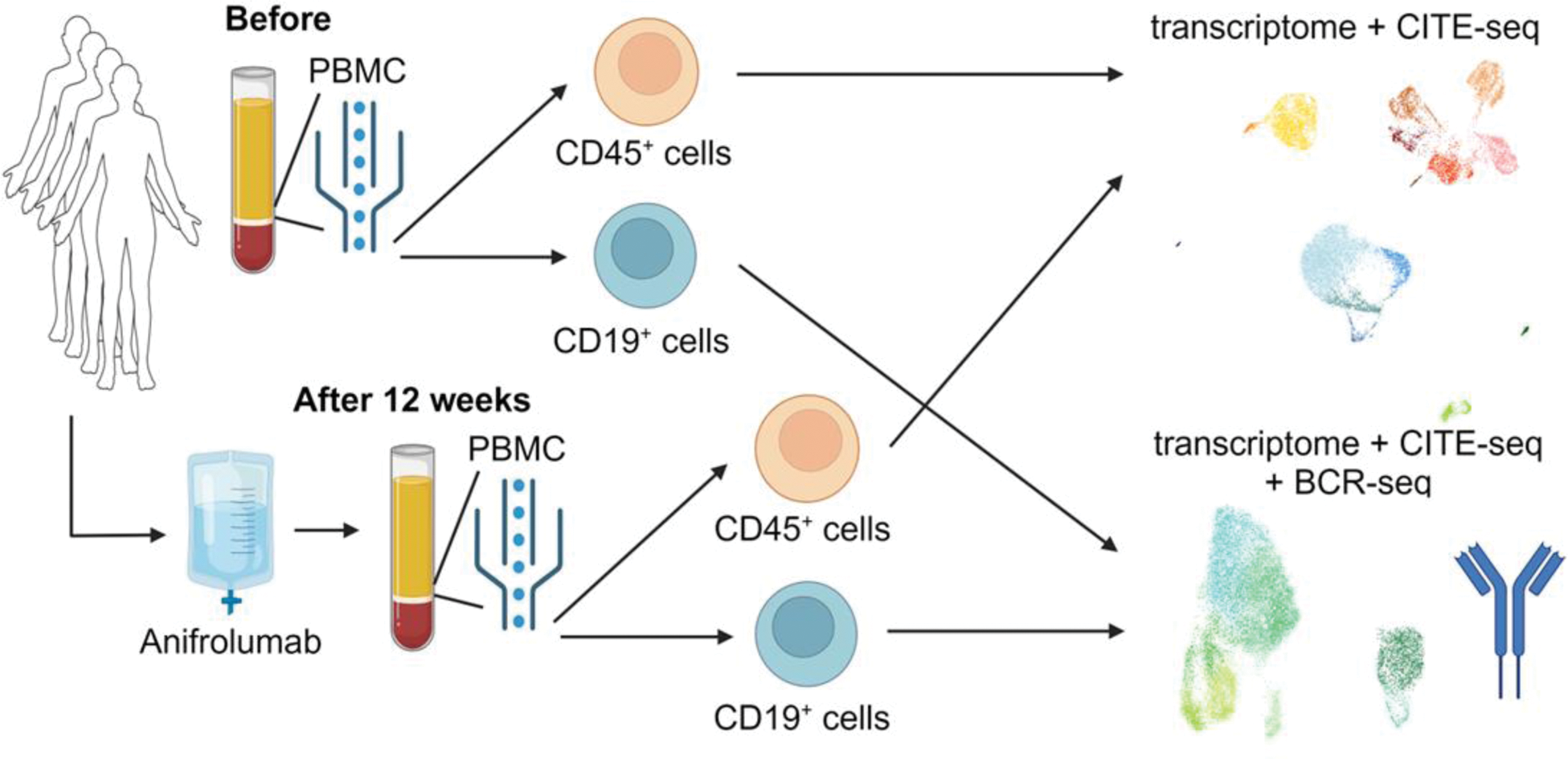
Methods: Peripheral blood mononuclear cells (PBMC) were collected from four patients with SLE at the University of Tokyo Hospital before and after 12 weeks of ANI treatment. Clinical data were obtained from medical records. Disease activity was assessed using SLE Disease Activity Index 2000 (SLEDAI-2K) scores, and glucocorticoid doses before and after 12 weeks of ANI treatment were recorded. CD45 + and CD19 + cells were sorted from PBMC, and single-cell sequencing was performed. For CD45 + cells, transcriptome and cellular indexing of transcriptomes and epitopes by sequencing analyses were conducted, and for CD19 + cells, BCR sequencing (BCR-seq) analysis was performed in addition to transcriptome and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) analyses (Figure 1). The ISG score was calculated as the mean z-scores of 21 ISGs [1]. V(D)J analyses for CD19 + B cells were performed using scRepertoire (v2.0.4) [2] and the Immcantation framework [3]. Statistical comparisons were conducted using Wilcoxon signed-rank tests.
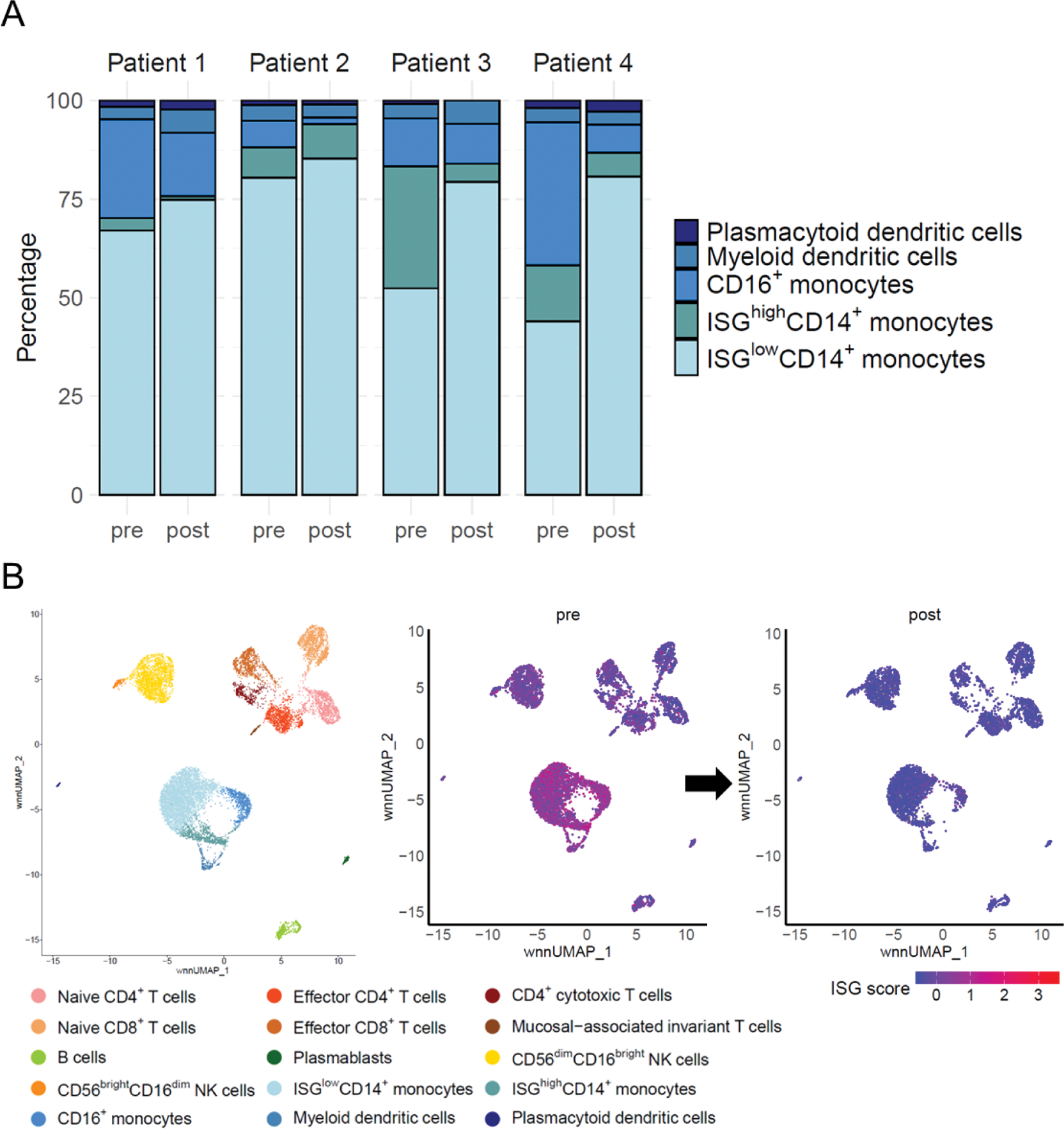
Results: All four patients were women with a mean age of 42.5 ± 6.0 years. The average disease duration was 16.2 ± 9.3 years, and the mean prednisolone dosage was 5.5 ± 3.1 mg at ANI initiation. Within 12 weeks, all patients showed a reduction in SLEDAI-2K scores or were able to taper glucocorticoids. Analysis of CD45 + cells showed a trend toward reduction in the proportions of ISG high CD14 + monocytes and CD16 + monocytes, populations with high ISG expression, among myeloid cells (Figure 2A). ISG score decreased significantly across all cell types (FDR-corrected p < 0.01, Figure 2B). Among CD19 + cells, there was a trend towards a decrease in the proportion of unswitched memory (USM) B cells. This was accompanied by an increase in the somatic hypermutation (SHM) ratio within USM B cells. ISG score significantly decreased across all B-cell subpopulations (FDR-corrected p < 0.01).
Conclusion: ANI treatment led to significant reductions in ISG scores across all cell subpopulations. Notably, there was a trend toward a decrease in subpopulation with high ISG expression, ISG high CD14 + monocytes and CD16 + monocytes, among the myeloid cells. Additionally, USM B cell proportions decreased, accompanied by an increase in SHM. The effects of ANI were evident at the cellular level within 12 weeks, and further studies with additional cases are needed to evaluate those changes in greater detail.
Overview of the Study. Peripheral blood mononuclear cells (PBMC) were collected before and 12 weeks after anifrolumab treatment from four patients with systemic lupus erythematosus (SLE). CD45 + and CD19 + cells sorted, and single cell RNA sequencing analysis and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) analyses were performed. For CD19 + cells, B cell receptor (BCR) sequencing (BCR-seq) was also performed.
Changes in CD45 + Cells Following Anifrolumab Treatment. (A) Changes in cell proportions among myeloid cells. (B) Alterations in interferon-stimulated gene (ISG) score in each subpopulation.
REFERENCES: [1] Yao Y et al. Development of Potential Pharmacodynamic and Diagnostic Markers for Anti-IFN-α Monoclonal Antibody Trials in Systemic Lupus Erythematosus. Hum Genomics Proteomics 2009:2009:374312.
[2] Borcherding N et al. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 2020:9:47.
[3]
Acknowledgements: This study was supported by AstraZeneca.
Disclosure of Interests: Keigo Terada: None declared, Yumi Tsuchida AstraZeneca, AbbVie, Janssen, GlaxoSmithKline, Astellas, Daiichi-Sankyo, Eli Lilly, Novartis, Eisai, Asahi Kasei, and AstraZeneca, Toshiyuki S Ushijima: None declared, Yuichi Suwa: None declared, Kazusa Saegusa: None declared, Saeko Yamada: None declared, Tomohisa Okamura Asahi Kasei, and Bristol-myers Squibb, Chugai, Keishi Fujio AstraZeneca, and Asahikasei Pharma, Chugai, Abbvie, Asahikasei Pharma, Bristol Myers, AstraZeneca, Tanabe Mitsubishi, Eisai, Gilead, Eli Lilly, Pfizer, Taisho, Astellas, Daiichi-Sankyo, Novartis, GlaxoSmithKline, and Alexion Pharma, Asahikasei Pharma, Chugai, Abbvie, Asahikasei Pharma, Bristol Myers, AstraZeneca, Eisai, Tsumura, and Taisho.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (