

Background: Juvenile idiopathic arthritis (JIA) results from an interplay of genetic and environmental factors, but the underlying mechanisms remain largely unknown. Familial, twin, and genome-wide association studies (GWAS) have helped to approach and dissect the genetic contribution to JIA [1, 2]. However, although JIA is more prevalent in girls and prevalences of disease phenotypes differ between the sexes, the role of sex-specific genetic associations in the disease has not been fully studied. Recently, we developed a polygenic risk score (PRS) for the children in the Norwegian Mother, Father and Child cohort study (MoBa) based on results from the largest GWAS of JIA to date, which included 3305 cases and 9196 controls [3, 4]. Understanding how the PRS associates with JIA and whether it performs similarly across sexes is essential for its utility in future studies.
Objectives: The aims of this study were 1) to investigate the relationship between the PRS for JIA and the probability of a JIA diagnosis, and 2) to explore whether the relationship between the PRS and JIA risk is different between males and females.
Methods: We studied the relationship between a PRS for JIA and a diagnosis of JIA in children from a large, prospective pregnancy cohort (MoBa) comprising approximately 114,500 children, 95,200 mothers, and 75,200 fathers [5]. We defined a JIA case as having at least two International Classification of Diseases (ICD)-10 codes (≥1 M08, ≥2 M09, or ≥1 M08 and ≥1 M09) registered in the Norwegian Patient Registry, a definition which was recently validated with a positive predictive value of 93.4% (4). The genotyping, quality control and imputation of the genetic data of the samples in MoBa have been extensively described previously [6]. We applied PRSice, version 2.3.3 [7] to calculate PRSs from the results of a previously published GWAS of JIA [3]. The first principal component (PC) was extracted for PRSs calculated across a range of different p-value thresholds, using this first PRS-PC as our final PRS for JIA. We standardized the PRS to a mean of zero and a standard deviation (SD) of 1 and used this standardized PRS for all analyses. Generalized linear models (GLM) and generalized additive models (GAM) were used within a logistic regression framework to assess the association. The top 10 PCs from the whole genotype dataset, sex, and year of birth were included as covariates in the models. Furthermore, we investigated whether the relationship between PRS and JIA differed by sex by applying GAM models with interaction terms.
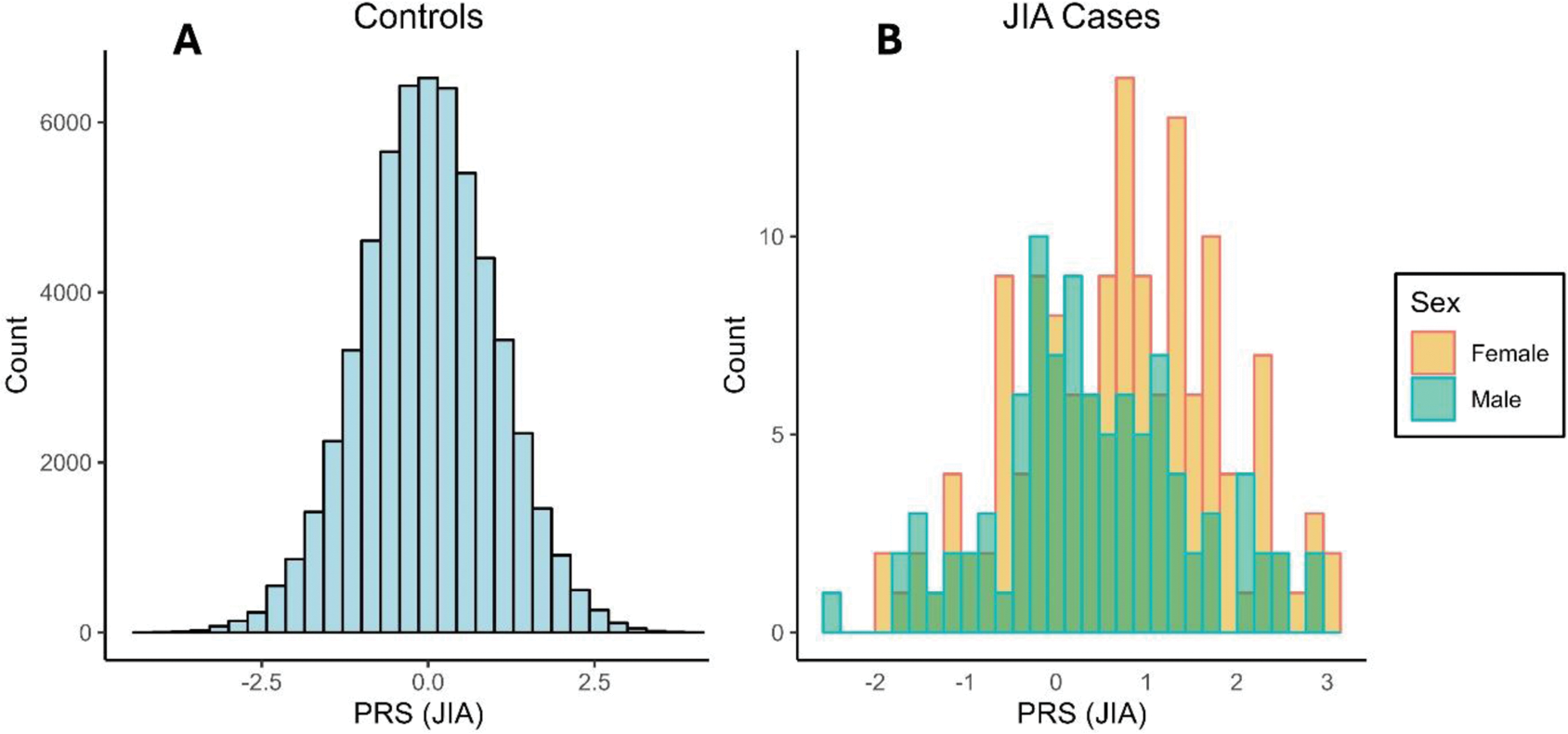
Results: Our final analytical sample included 238 JIA cases (91 males and 147 females) and 57,392 controls. PRS was significantly associated with JIA using both GLM (p < 2e-16) and GAM (p < 2e-16) models, and our results indicated a nonlinear relationship between PRS and JIA (effective degrees of freedom, EDF = 1.96). The PRS mean was 0.40 for male JIA cases and 0.70 for female JIA cases, indicating a stronger association between PRS and JIA diagnosis in females compared to males (Figure 1). PRS was significantly associated with JIA in both females (p < 2e-16) and males (p < 0.001). We found a significant interaction between sex and JIA PRS in relation to JIA (p = 0.017), and indications of a stronger and more logit-nonlinear relationship in females (EDF = 1.82) versus males (EDF = 1.06) (Figure 2).
Distribution of JIA PRS in (A) controls, and (B) JIA cases grouped by sex
Relationship between PRS of JIA and probability of JIA in females and males. The lines show the fitted prediction model of JIA probability ~ PRS + sex + year of birth + top 10 principal components for each of the sexes. The colored areas represent the 95% confidence intervals for the corresponding models.
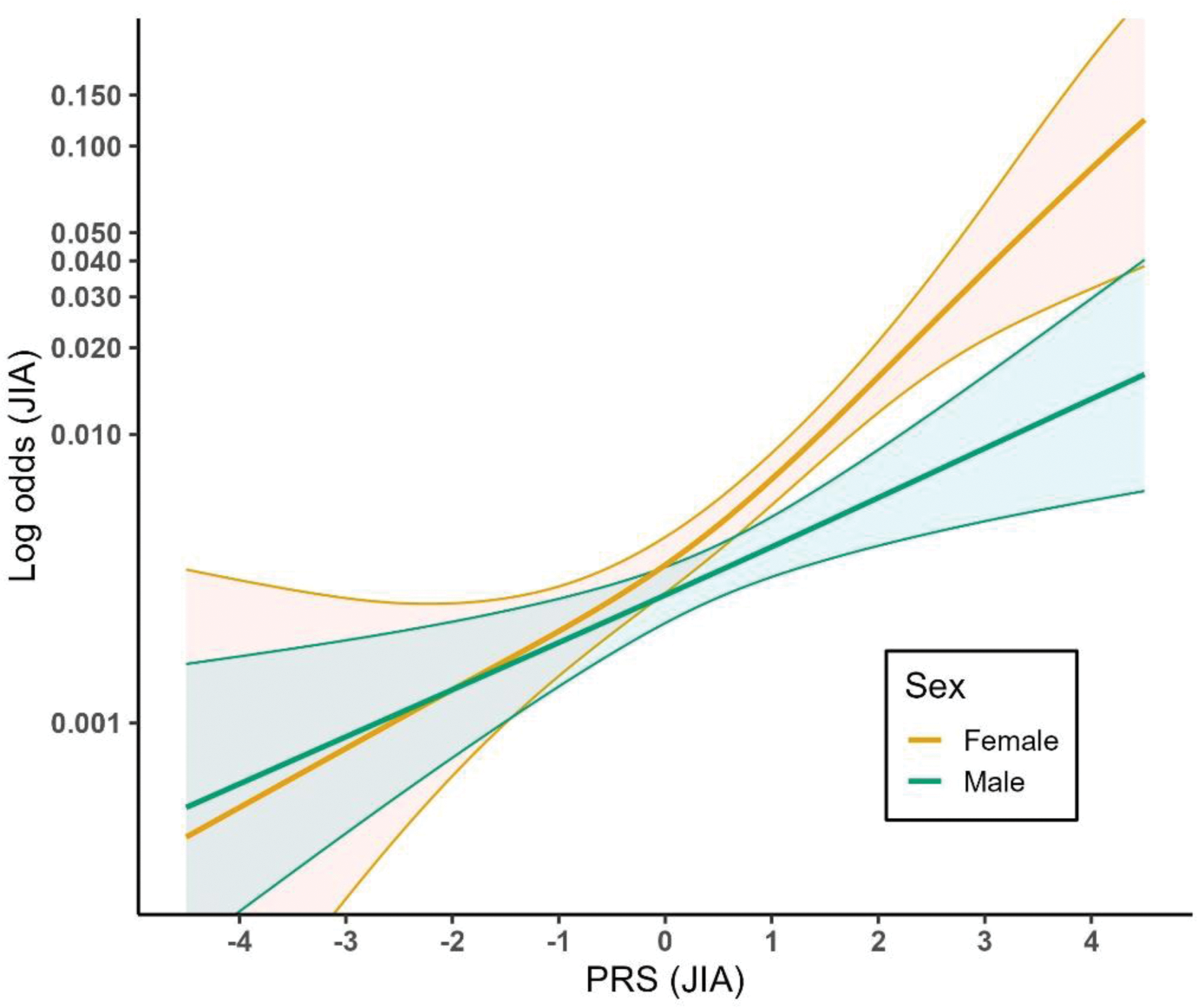
Conclusion: The relationship between PRS and JIA was slightly logit-nonlinear, but only for females. The PRS for JIA can be useful in analyses, but sex-stratification is recommended. Future studies should further investigate sex differences in genetic predisposition to JIA and other autoimmune diseases.
REFERENCES: [1] La Bella S., et al. Int J Mol Sci. 2023;24(3).
[2] Cobb JE., et al. Rheumatology (Oxford). 2014;53(4):592-9.
[3] López-Isac E., et al. Ann Rheum Dis. 2021;80(3):321-8.
[4] Hestetun SV., et al. Arthritis Rheumatol. 2024.
[5] Magnus P. et al. Int J Epidemiol. 2016;45(2):382-8.
[6] Corfield EC., et al. bioRxiv. 2024:2022.06.23.496289.
[7] Choi SW., et al. Gigascience. 2019;8(7).
Acknowledgements: We are grateful to all the participating families in MoBa, and we thank the Norwegian Institute of Public Health (NIPH) for generating high-quality genomic data.
Disclosure of Interests: Kristine Løkås Haftorn: None declared, Hamid Khoshfekr Rudsari: None declared, Piotr Pawel Jaholkowski: None declared, Vilde Øverlien Dåstøl: None declared, Sigrid Valen Hestetun: None declared, Ole A. Andreassen has received speaker fees from Lundbeck, Janssen, Otsuka, and Sunovion, has received consulting fees from Cortechs.ai and Precision Health AS, Clarice R. Weinberg: None declared, Helga Sanner: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (