

Background: Juvenile Idiopathic Arthritis (JIA) is the most common form of chronic arthritis in childhood. The adaptive branch of the immune system plays a crucial role in the development of JIA. An hyperactivation of B cells characterizes patients with oligoarticular- and polyarticular- (oligo-/poly-) JIA (Barnes MG, A&R 2010) and we have shown that switched memory B cells are expanded in this group of patients (Marasco E, A&R 2018). Preliminary data from our group demonstrated that B cells of oligo-/poly-JIA patients exhibit changes in the immunoglobulin repertoire, including an altered peripheral selection process and lower frequency of somatic hypermutation, suggesting an extrafollicular activation of autoreactive B cells (Marasco E, EULAR Abstract 2024).
Objectives: Building on our previous work, we aimed to investigate the signalosome and transcriptomic profile of B cells in patients with oligo-/poly-JIA.
Methods: We enrolled patients with a diagnosis of oligo-/poly-JIA (n=15) and age matched controls (n=15). Intracellular levels of phospho-proteins (Syk, BLNK, ERK1-2, BTK, PLCγ) and surface levels of CD22 were evaluated by flow cytometry. Calcium mobilization after anti-Ig stimulation was evaluated with Indo-1 by flow cytometry. Transcriptomic analysis was performed on sorted naïve B cells of patients and controls activated with anti-Ig, rhCD40L and IL-21 (n=4 for each group). Total RNA was extracted with Trizol. Libraries were prepared with SureSelect XT HS2 RNA System (Agilent), obtaining a strand-specific library (anti-sense strand for the forward read) positively enriched for human exons (SureSelect XT HS Human All Exon V8 Probe), following the SOPs. Samples were run on a NextSeq 500/550 (Illumina) with a 2 x 101 bp sequencing strategy.
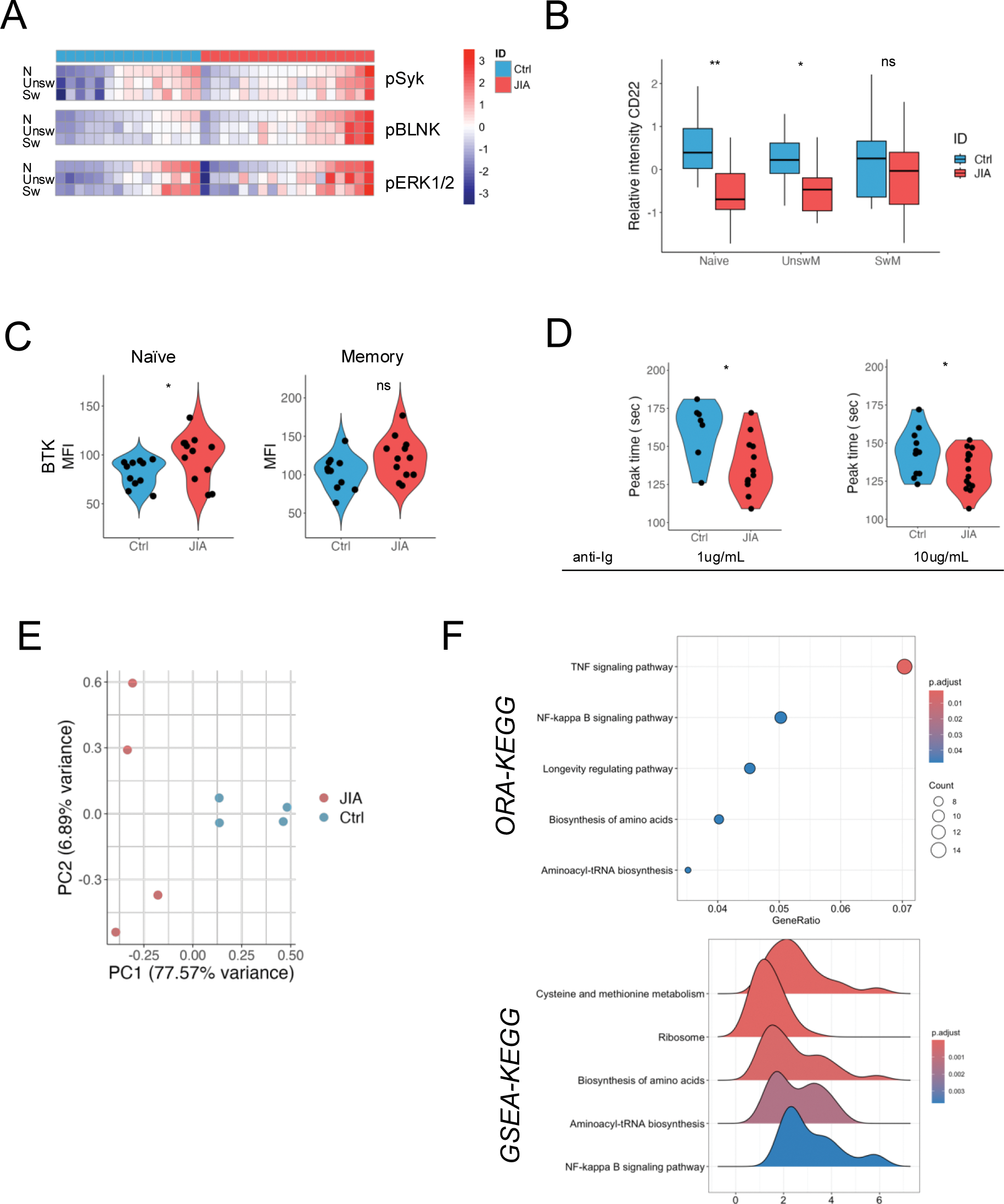
Results: We first analyzed the basal expression and phosphorylation levels of key proteins involved in the regulation of B-cell receptor (BCR) signaling (Syk, BLNK, ERK1/2) in B cell of oligo-/poly-JIA patients and controls. We observed a higher frequency of phosphorylation of BLNK, but not of Syk and ERK1/2 in both naïve and memory B cells compared to controls (Figure 1A). Next, we evaluated the surface expression of CD22, a negative regulator of B cell signaling in all B cell subsets: we observed a lower surface expression of CD22 in naïve B cells and unswitched memory (UnswM), but not in switched memory B cells (SwM) (Figure 1B), suggesting that most alterations in BCR signaling are established at the mature naïve stage. Upon activation with an anti-Ig, naïve B cells showed higher phosphorylation of BTK but not PLCγ (Figure 1C). As BLNK, CD22 and BTK regulate the calcium mobilization following BCR stimulation, we investigated calcium mobilization after BCR activation. Upon in vitro stimulation of BCR with anti-Ig, we observed that naïve B cells of patients with JIA showed a faster calcium response to BCR activation, with a shorter peak time compared to controls at two different concentrations of anti-Ig (Figure 1D). We focused on naïve B cells and investigated the transcriptomic profile of activated naïve B cells. After quality controls, gene annotation to the reference genome, counting and filtering for low expression genes, we identified 13161 genes. We performed a PCA analysis: the first 2 components explained 84.5% of the total variance and the two groups of samples separated on the PCA plot, indicating differences in gene expression between the two groups (Figure 1E). With a significance threshold of adjusted p-value set at 0.05, we identified 372 differentially regulated genes (DEGs) between patients and controls with the algorithm DESeq2. We performed on our set of DEGs over-representation analysis (ORA) and gene set enrichment analysis (GSEA). Both analyses confirmed that most of the DEGs were related to immune signaling of the TNF family and of NF-kB, and genes involved in the metabolisms of amino acids and protein synthesis (Figure 1F). We also identified two differentially regulated long non coding RNAs (lnRNAs) with known functions: ENSG00000256482 plays a role in regulating amino acid metabolisms, while ENSG00000262151 is an antisense to CIITA, a gene that is important for antigen presentation by B cells.
Conclusion: Our data show that naïve B cells of JIA patients exhibit an increased phosphorylation of BLNK together with a reduced expression of CD22. Upon activation of BCR, naïve B cells respond more promptly to BCR activation with faster calcium mobilization. Transcriptomic analysis of activated naïve B cells revealed differential expression of genes associated with TNF family immune signaling, NF-kB pathways, antigen presentation, and amino acid metabolism, as well as those involved in protein synthesis. These latter pathways are regulated by mTOR, a critical mediator of differentiation toward the extrafollicular pathway in B cells, highlighting a potential link between B cell activation outcomes and metabolism in JIA, warranting further investigation.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: Emiliano Marasco BMS, Angela Aquilani: None declared, Giusyda Tarantino: None declared, Maria Isabella Petrone: None declared, Rebecca Nicolai: None declared, Ivan Caiello: None declared, Sabina Barresi: None declared, Silvia Magni-Manzoni: None declared, Fabrizio De Benedetti SOBI, NOVARTIS.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (