

Background: The employment of targeted gene panels for Next-Generation Sequencing (NGS) is a cornerstone in supporting the diagnosis of systemic autoinflammatory diseases, mostly when the clinical phenotype seems unclear.
Objectives: This study aims to identify mutations and variants of unknown significance (VUS) to determine whether patients presenting with autoinflammatory clinical features can be categorised into specific subgroups. Additionally, it explores whether VUS may correlate with distinct clinical phenotypes, transitioning from the unspecific “Systemic undifferentiated recurrent fevers (SURF)” paradigm toward new potential sAID subgroups.
Methods: Patients with clinical and biochemical inflammatory features, referred to the outpatient Clinic for Autoinflammatory Diseases of Padova University Hospital, were retrospectively reviewed from February 2023 to November 2024. NGS sequencing was performed using Custom “Fever & Autoinflammatory Disease” panel (SOPHIA Genetics) in Illumina MiSeq, which analyzes coding regions of 17 genes ( ADA2, CARD14, ELANE, IL10RB, IL10RB, IL1RN, LPIN2, MEFV, MKV, NLRP12, NLRP3, NLRP7, NOD2, PSMB8, PSTPIP1, TNFRSF11A, TNFRSF1A ). Variant calling and data analysis were performed by the Sophia-DDM-V6.5 bioinformatics analysis programme. The interpretation of the variants was performed according to the 2015 ACMG standards and guidelines.
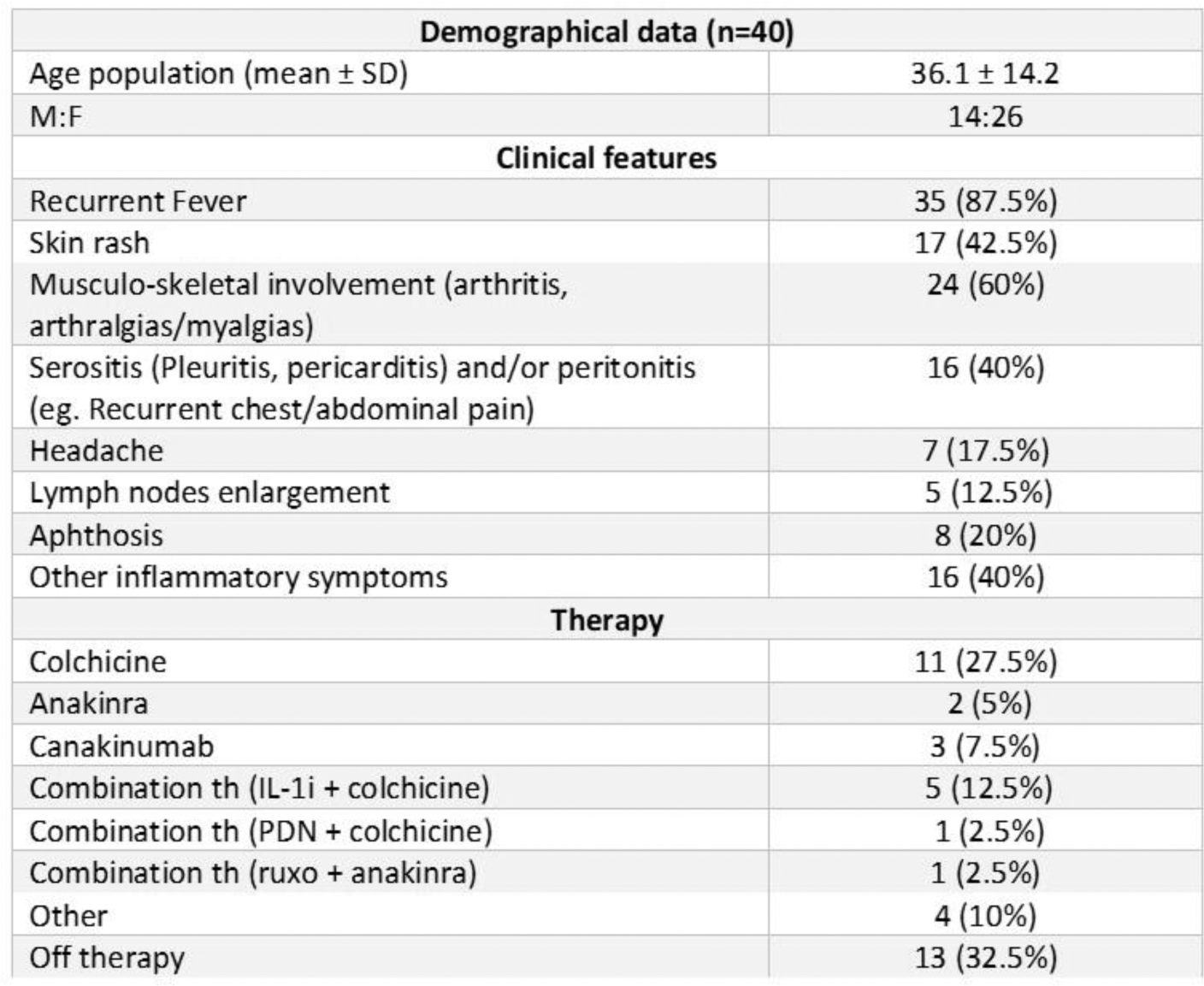
Results: Among 103 patients screened, 80 (77.7%) displayed at least one retained non-synonymous variant with a minor allele frequency (MAF)≤0.05. Of these, 34 carried at least one VUS, and 6 had pathogenic variants confirming the phenotype. Clinical and demographical data are shown in Figure 1. Amongst variations of interest, two distinct clusters of patients were identified with VUS on NOD2: one group with p.Arg702Trp predominantly presenting with recurrent episodes of hyperpyrexia and one with p.Leu1007Profs*2 primarily exhibiting oral aphthosis and cutaneous rashes. On NRLP12 four patients had the p.F402L variant exhibiting heterogeneous symptoms and severity; finally, patients carrying two VUS across the same or different genes generally experienced a higher burden of inflammatory symptoms and responded well to colchicine or IL-1 inhibitors.
Conclusion: A validated gene panel for NGS is pivotal for identifying mutations that confirm specific clinical phenotypes. Although patients with VUS remain classified as “SURF,” emerging clusters of patients sharing the same VUS and clinical profiles suggest the possibility of “novel” gene-related pathologies. Further investigations, including Whole-Exome Sequencing or functional studies, will be essential for enhancing the diagnostic framework for VUS carriers.
REFERENCES: NIL.
Clinical and demographical data of the patients carrying VUS
Acknowledgements: NIL.
Disclosure of Interests: Sara Bindoli: None declared, Anna Fu: None declared, Stefania Moz: None declared, Andrea Doria GSK, Otsuka, Eli Lilly, Paolo Sfriso SOBI, Novartis, Paola Galozzi: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (