

Background: Familial Mediterranean Fever (FMF) is the most common hereditary autoinflammatory syndrome, and colchicine resistance remains a significant challenge in its treatment. The ability to predict colchicine resistance in advance could greatly impact treatment management. Recently, machine learning algorithms have emerged as powerful tools for predicting this resistance. These algorithms are capable of forecasting colchicine resistance based on various patient features, enhancing clinical decision-making.
Objectives: To enhance the performance of baseline models by incorporating new features and technically improving the models.
Methods: In this study, a cohort of 965 patients was examined, with various clinical and genetic features used as inputs for the models. The patients with missing data on new features were eliminated. The features included mutation type (homozygous, heterozygous, compound heterozygous), presence of specific mutations (M694V, M680I, E148Q, V726A, R761H), presence of recurrent arthritis, presence of arthralgia, presence of oligoarthritis, age of diagnosis and frequency of attacks. Two machine learning techniques were employed: Deep Learning and Logistic Regression. These models were trained on the dataset to predict colchicine resistance based on the newly introduced features.
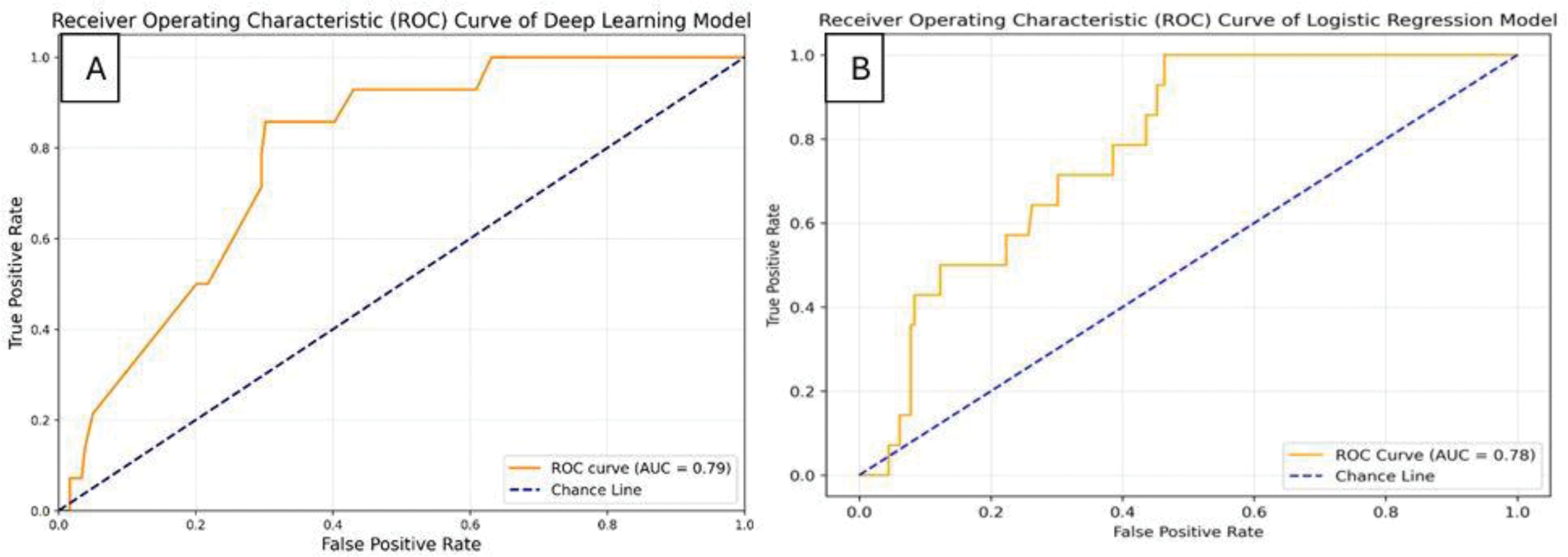
Results: The deep learning model achieved an AUC of 0.79, while the logistic regression model reached an AUC of 0.78 (Figure 1). These results were compared with baseline models, and the comparison is shown in Table 1.
ROC Curves of Machine Learning Models
Comparison of Baseline and Current Machine Learning Models
| Model | Features (Baseline Model) | Features (Current Model) | AUC
| AUC (Current Model) |
|---|---|---|---|---|
| Logistic Regression | Presence of compound heterozygous mutations in the MEFV gene,
| Mutation type (Homozygous, heterozygous, compound heterozygous),
| 0.76 | 0.78 |
| Deep Neural Network | Number of attacks per month before treatment,
| Mutation type (Homozygous, heterozygous, compound heterozygous),
| 0.75 | 0.79 |
Conclusion: The incorporation of new features and technical enhancements has led to significant improvements in the predictive performance of our models. These models show a promising ability to predict colchicine resistance more accurately and can be further refined for even better performance.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (