

Background: Accumulating data supports that glucose dysregulation increases the risk of immune mediated inflammatory disease (IMIDs), the causal associations between glucose-lowering drug and IMIDs remain inconclusive.
Objectives: The study aimed to investigate whether genes encoding glucose-lowering drug targets are causally associated with the risk of IMIDs.
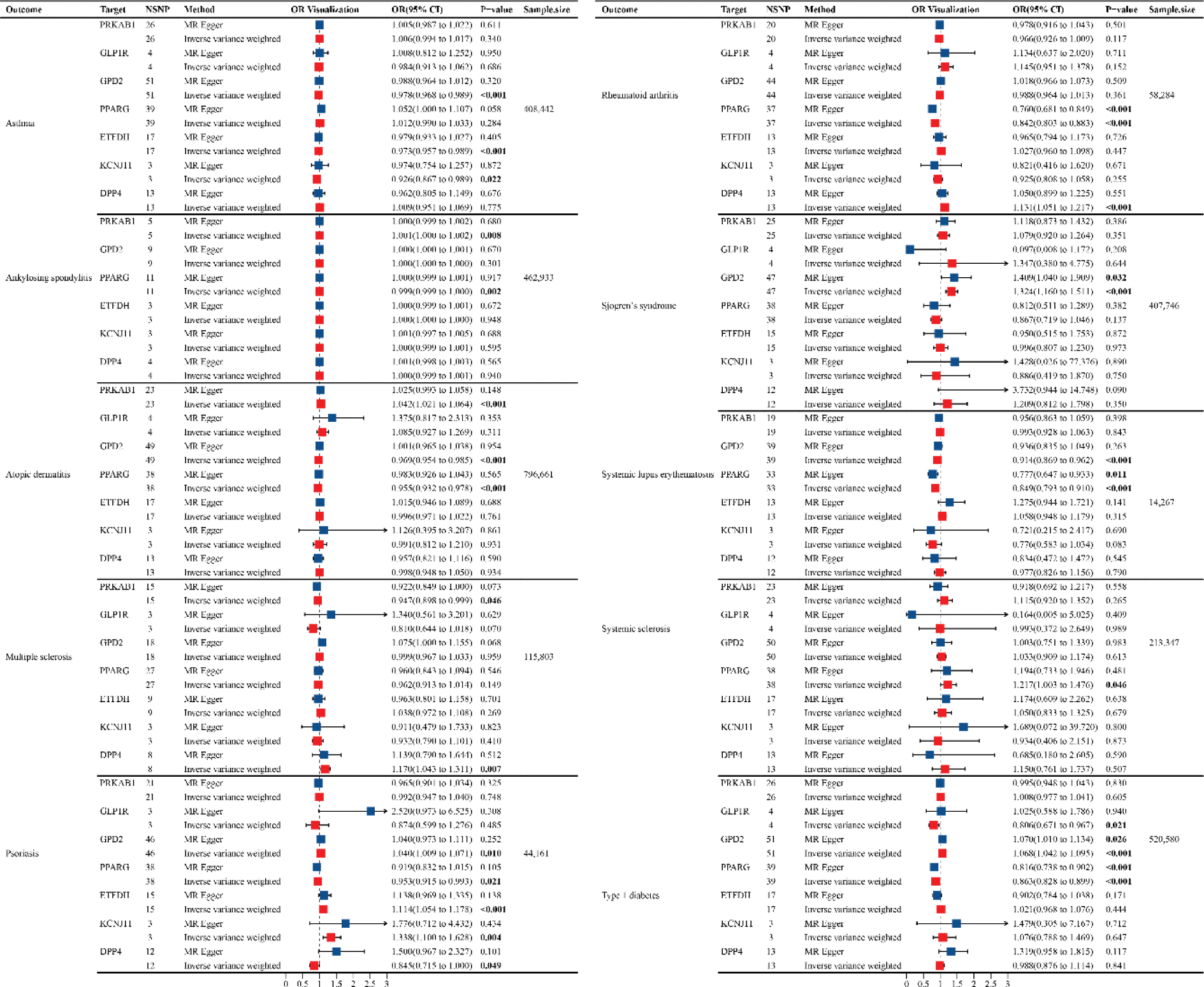
Methods: We developed genetic instruments for glucose-lowering drug targets (metformin [PRKAB1, ETFDH, GPD2], thiazolidinediones [PPARG], sulfonylureas [KCNJ11], glucagon-like peptide 1 analogues [GLP1R]; dipeptidyl peptidase 4 inhibitor [DPP4]) using summary genome-wide association data. Genetic instruments were constructed using cis-acting genome-wide significant (p<5×10-5) single nucleotide polymorphisms (SNPs) permitted to be in linkage disequilibrium (r2<0.001). Summary genetic association estimates for these SNPs were obtained from genome-wide association study (GWAS) consortia for the following IMIDs: asthma, ankylosing spondylitis, atopic dermatitis, multiple sclerosis, psoriasis, rheumatoid arthritis, Sjogren’s syndrome, systemic lupus erythematosus, type 1 diabetes, systemic sclerosis, inflammatory bowel disease. Inverse-variance weighted random-effects models adjusting for linkage disequilibrium were employed to estimate causal associations between genetically proxied drug target perturbation and risk of IMIDs. A colocalization posterior probability (PPH4) >80% was employed to indicate support for shared causal variants across drug targets and IMIDs.
Results: In MR analysis, genetically proxied PPARG was associated with a decreased risk of systemic lupus erythematosus (OR 0.85, 95% CI 0.79-0.91), rheumatoid arthritis (OR 0.84, 95% CI 0.80-0.88), type 1 diabetes (OR 0.86, 95% CI 0.83-0.90), atopic dermatitis (OR 0.96, 95%CI 0.93-0.98), psoriasis (OR 0.95, 95%CI 0.92-0.99); ETFDH was associated with decreased risk of asthma (OR 0.97, 95%CI 0.96-0.99); GLP1R was associated with a decreased risk of type 1 diabetes (OR 0.81, 95%CI 0.67-0.97); GPD2 was associated with a decreased risk of systemic lupus erythematosus (OR 0.91, 95%CI 0.87-0.96), asthma (OR 0.98, 95%CI 0.97-0.99), atopic dermatitis (OR 0.97, 95%CI 0.95-0.99); PRKAB1 was associated with a decreased risk of multiple sclerosis (OR 0.95, 95%CI 0.90-0.99); KCNJ11 associated with a decreased risk of asthma (OR 0.93, 95%CI 0.87-0.99). In contrast, PPARG was associated with an increased risk of systemic sclerosis (OR 1.22, 95%CI 1.00-1.48); PRKAB1 was associated with an increased risk of atopic dermatitis (OR 1.04, 95%CI 1.02-1.06); KCNJ11(OR 1.34, 95%CI 1.10-1.63) and ETFDH (OR 1.11, 95%CI 1.05-1.18) associated with an increased risk of psoriasis; GPD2 was associated with an increased risk of type 1 diabetes (OR 1.07, 95%CI 1.04-1.10), Sjogren’s syndrome (OR 1.32, 95%CI 1.16-1.51), psoriasis (OR 1.04, 95%CI 1.01-1.07); DPP4 was associated with an increased risk of rheumatoid arthritis (OR 1.13, 95%CI 1.05-1.21). A strong colocalizations association with PPARG were observed for rheumatoid arthritis and (rs1699346, PP.H4 = 0.99).
Conclusion: The drug target MR analyses identified several glucose-lowering drug targets associated with IMIDs. These results may guide clinicians in the treatment of individuals at high risk of IMID. Future studies are warranted to clarify the underlying mechanistic pathways between glucose metabolism and risk of IMIDs.
REFERENCES: [1] Di Niu et al, Int Immunopharmacol. 2024 Mar 30:130:111682.
[2] Dunfang Zhang et al, Immunity. 2019 Oct 15;51(4):671-681.e5.
[3] Shiying Shao et al, Pharmacol Ther. 2022 Nov:239:108270.
[4] Lu Wang et al, Front Immunol. 2022 Sep 20:13:952398.
[5] Georg Schett et al, N Engl J Med. 2021 Aug 12;385(7):628-639.
[6] GBD 2019 IMID Collaborators et al, EClinicalMedicine. 2023 Sep 9:64:102193.
Acknowledgements: We thank the authors who prepared these publicly available data. This study is based on publicly available data.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (