

Background: The prevalence of gout, a chronic metabolic disease, has increased in recent years. Predicting gout in advance is very useful since it can prevent not only gout but also its complications. There have been various attempts to predict gout with clinical variables. The polygenic risk score (PRS) is a useful tool for predicting outcomes of this condition. However, the clinical utility of the PRS for disease prediction remains controversial.
Objectives: The purpose of this study is to evaluate whether the addition of the PRS to conventional risk factors could improve the performance of ML-based prediction model for gout.
Methods: Using data from the Korean Genome and Epidemiology Study, machine learning (ML) models were developed to predict gout based on the PRS and clinical variables, such as uric acid, lifestyle habits, and metabolic syndrome (MetS) profile. Five different ML algorithms were applied, including logistic regression, random forest (RF), decision tree, extreme gradient boosting, and light gradient boosting machine.
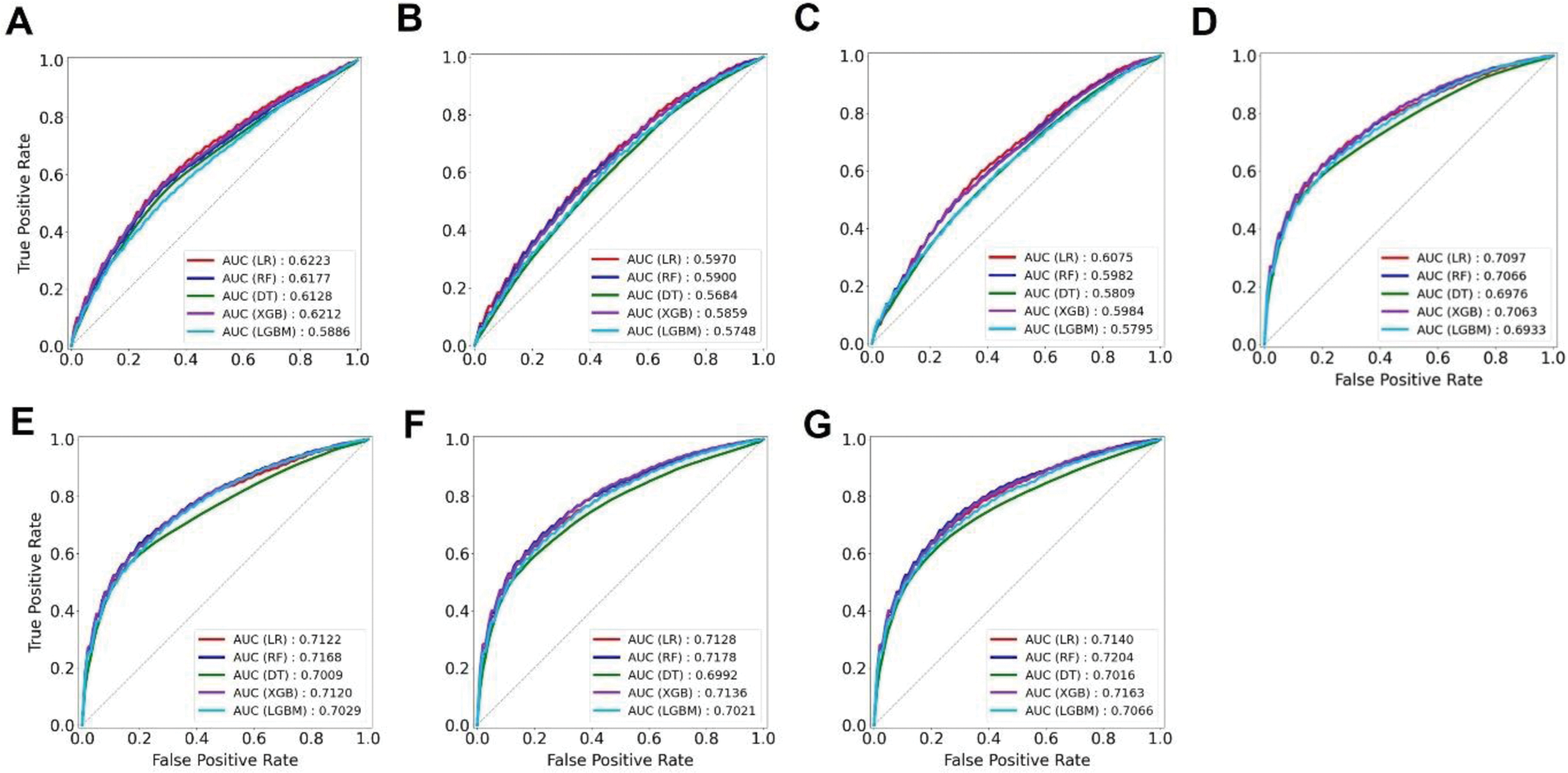
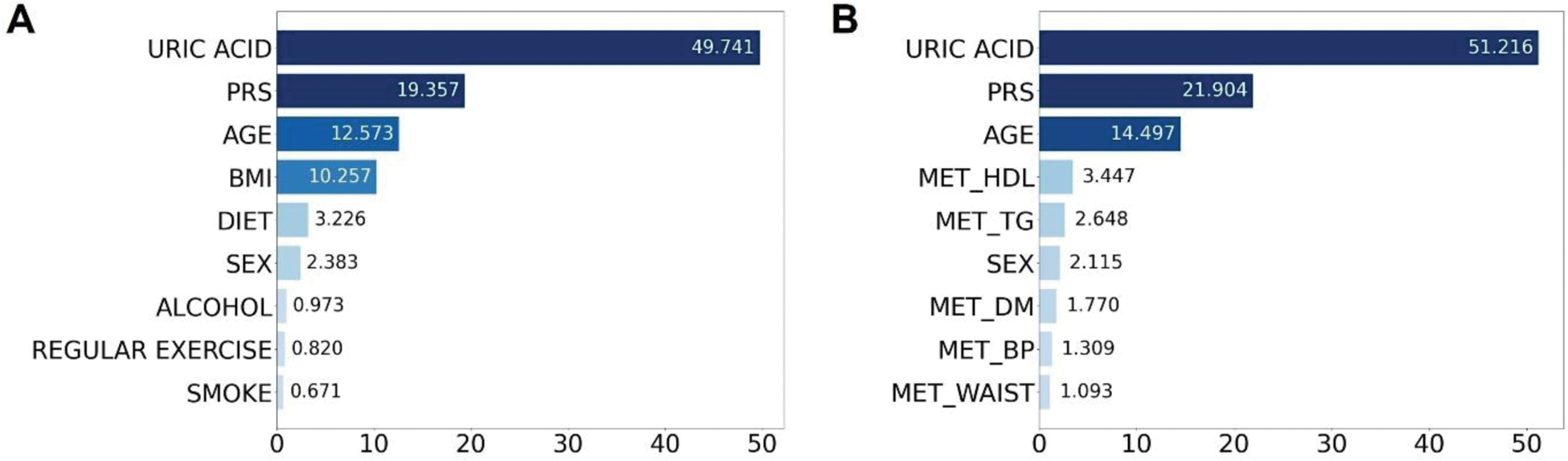
Results: Among five ML models, the RF model that incorporated PRS, age, sex, MetS, and uric acid showed the highest area under the curve value (0.7204, 95% CI = 0.7124-0.7284) (Figure 1). Feature importance analysis highlighted uric acid as the most important predictor of gout, followed by PRS and age (Figure 2). Although the PRS enhanced the predictive power of the ML models, its effect was modest, suggesting that traditional risk factors remain important for gout prediction.
Conclusion: We developed an ML-based prediction model using an Asian cohort and compared the performance of the clinical variable and PRS-based models. The addition of PRS to clinical variables enhanced the predictive power of the ML model for gout, though the effect of PRS was modest.
Receiver operating characteristic curves predicting gout. (A) PRS only; (B) age, sex, lifestyle habits; (C) age, sex, MetS; (D) age, sex, lifestyle habits, uric acid; (E) age, sex, MetS, uric acid; (F) age, sex, lifestyle habits, PRS, uric acid; and (G) age, sex, MetS, PRS, uric acid
Feature importance bar plot of RF classifier. Bar plot showing feature importance in the random forest model for two feature sets: (A) importance using age, sex, lifestyle habits, PRS, and uric acid; (B) importance using age, sex, metabolic syndrome, PRS, and uric acid. The numbers in the graph represent feature importance values, summed across 100 iteration models.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (