

Background: The intestinal microbiome plays a fundamental role in immune regulation and other physiological processes such as metabolism and vitamin production. Increasing evidence suggests its involvement in systemic lupus erythematosus (SLE) pathogenesis, progression, and symptom severity. However, existing research presents conflicting results due to factors such as diet, genetics, geographical location, and treatment regimens. Understanding the microbiome’s role in SLE may open new avenues for therapeutic interventions, such as microbiota modulation.
Objectives: The primary objective of this study was to determine the composition of the intestinal microbiome in SLE patients compared to a healthy control group. Secondary objectives included assessing differences in alpha and beta diversity, identifying taxonomic variations, evaluating the impact of immunosuppressive therapies on microbial diversity, and exploring the relationship between dietary habits, particularly adherence to the Mediterranean diet, and microbiome composition.
Methods: This observational case-control study included 30 participants (20 SLE patients with low disease activity and 10 healthy controls) recruited at a single center between September 2023 and January 2024. All participants provided informed consent, and the study was approved by the regional ethics committee. Stool samples were collected and processed for 16S rRNA sequencing using the Ion Torrent platform. Taxonomic distribution, diversity metrics, and operational taxonomic units (OTUs) were analyzed with QIIME2 software. Clinical and dietary data were collected using standardized questionnaires. Disease activity and cumulative damage were evaluated with the SLEDAI-2K and SLICC indices, respectively. Statistical analyses were performed to compare microbiota composition and diversity between groups and assess correlations with clinical and treatment variables.
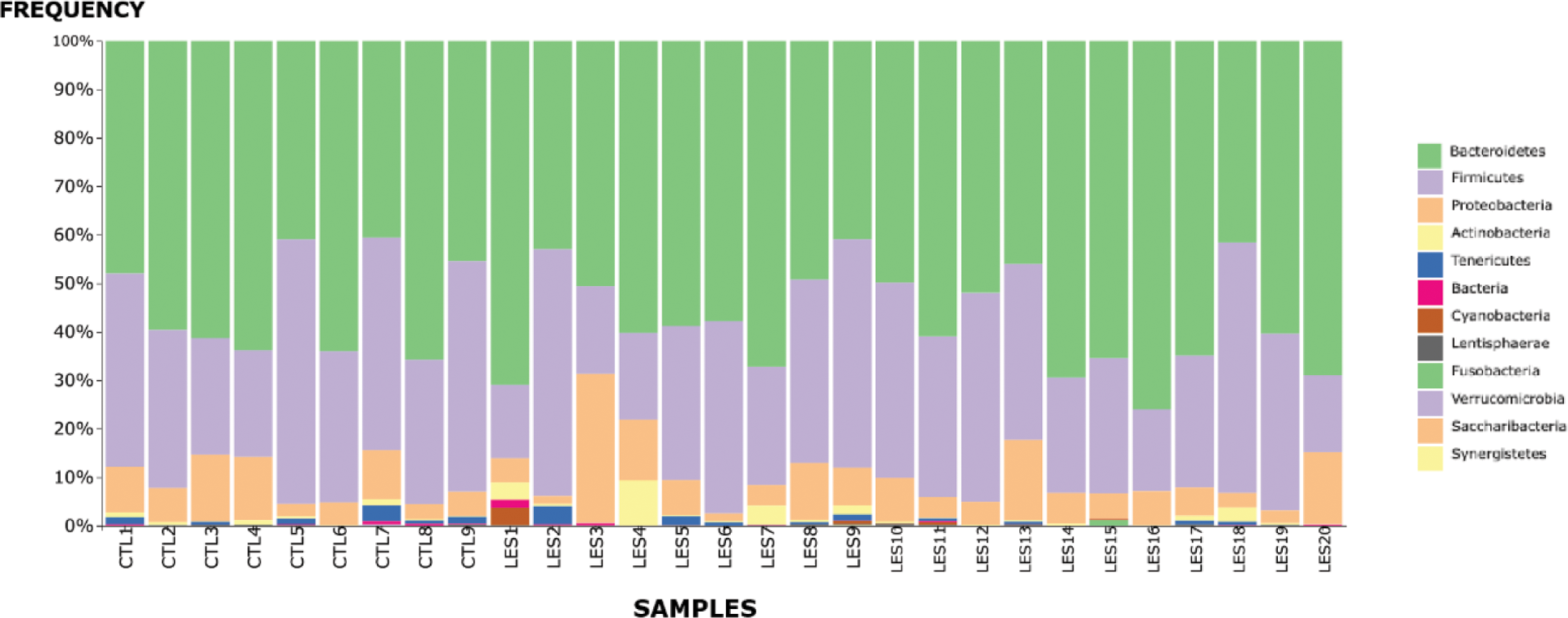
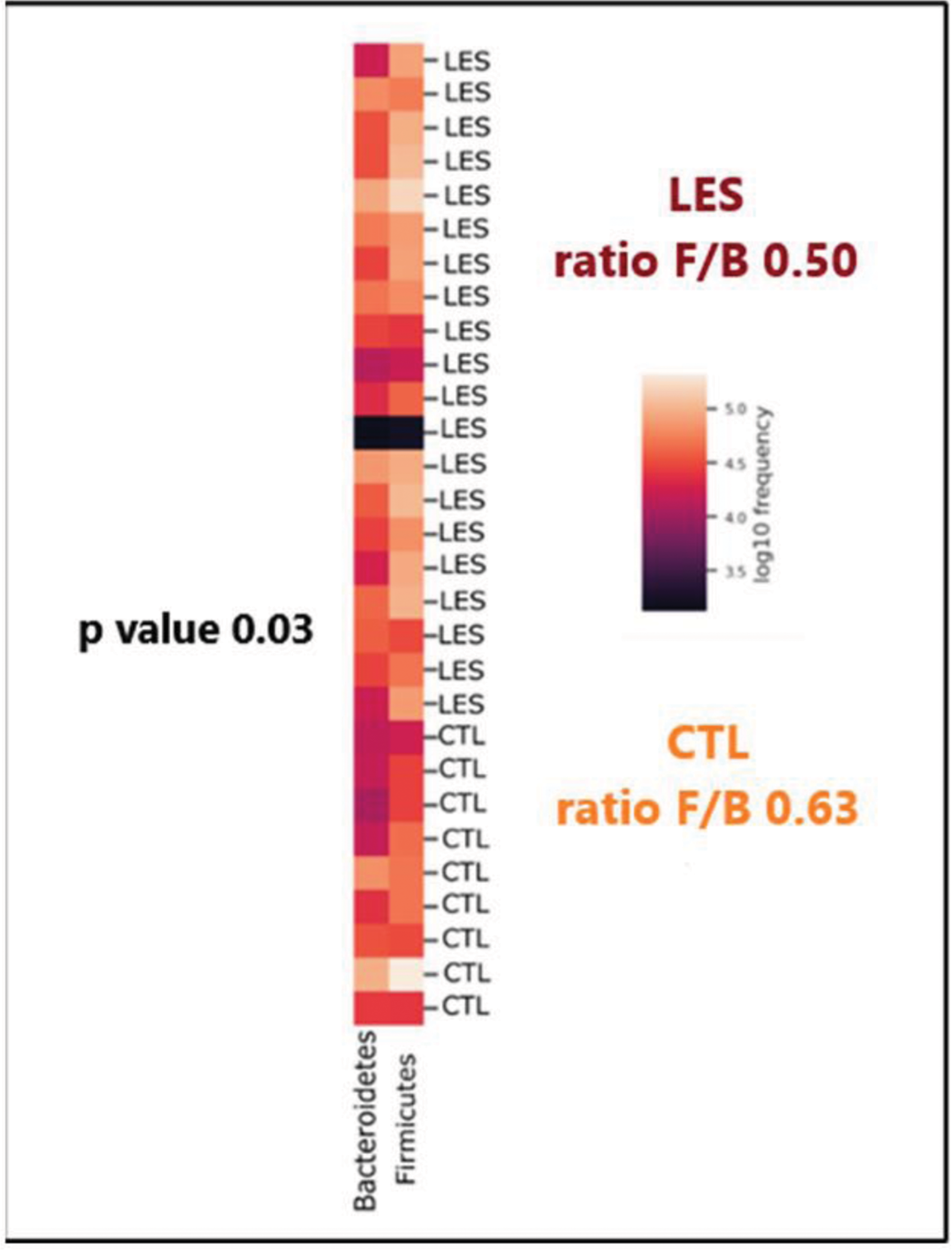
Results: The study population included 20 female SLE patients (mean age: 49.8 years) and 10 matched controls. SLE patients exhibited significantly lower microbial uniformity (Pielou’s evenness, p = 0.03) compared to controls, while other alpha and beta diversity metrics showed no significant differences. Subgroup analyses revealed reduced microbial diversity in patients treated with low-dose glucocorticoids (≤7.5 mg/day), with statistically significant reductions in Butyricimonas and Lachnospiraceae abundance. Taxonomic analysis demonstrated a decreased Firmicutes/Bacteroidetes ratio (p = 0.03) in SLE patients. Additionally, increased abundances of Rikenellaceae, Ruminococcaceae, and Enterobacteriaceae families were observed alongside reductions in Parabacteroides distasonis and Prevotella copri. Notably, patients with lupus nephritis (n=2) exhibited elevated levels of Desulfovibrio and Ruminococcaceae. Dietary adherence to the Mediterranean diet was not significantly associated with diversity indices, but patients adhering to the diet displayed trends toward greater microbial diversity. No significant differences in beta diversity were observed, indicating relatively uniform community composition between groups.
Conclusion: This study highlights significant alterations in the intestinal microbiome of SLE patients, including reduced microbial diversity and distinct taxonomic shifts. These findings emphasize the potential role of dysbiosis in SLE pathogenesis and its modulation by glucocorticoid therapy. While the Mediterranean diet’s impact on microbial diversity requires further exploration, the observed trends suggest potential benefits. Future research with larger cohorts is necessary to validate these observations and assess microbiota-targeted interventions as therapeutic options for SLE.
Taxonomic distribution
Firmicutes/Bacteroidetes ratio
REFERENCES: [1] Xiang S, Qu Y, Qian S, et al. Association between systemic lupus erythematosus and disruption of gut microbiota: a meta-analysis. Lupus Sci Med. 2022;9(1):e000599.
[2] Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857.
[3] Hevia A, Milani C, López P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5(5):e01548-14.
[4] Huang C, Yi P, Zhu M, et al. Safety and efficacy of fecal microbiota transplantation for treatment of systemic lupus erythematosus: An EXPLORER trial. J Autoimmun. 2022;130:102844.
[5] Wang M, Zhu Z, Lin X, et al. Gut microbiota mediated the therapeutic efficacies and the side effects of prednisone in the treatment of MRL/lpr mice. Arthritis Res Ther. 2021;23(1):240.
[6] Zhao M, Wen X, Liu R, et al. Microbial dysbiosis in systemic lupus erythematosus: a scientometric study. Front Microbiol. 2024;15:1319654.
[7] Pan Q, Guo F, Huang Y, et al. Gut Microbiota Dysbiosis in Systemic Lupus Erythematosus: Novel Insights into Mechanisms and Promising Therapeutic Strategies. Front Immunol. 2021;12:799788.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (