

Background: Septic arthritis is a medical emergency, which if not appropriately and timely treated, presents with joint destruction, sepsis, and death [1]. Pediatric septic arthritis (PSA) presents one of the biggest diagnostic challenges because of the subtlety of clinical symptoms and laboratory tests lacking sensitivity [2]. Staphylococcus aureus is the most common and one of the most aggressive causative microorganisms of prosthetic shoulder arthroplasty, accounting for a high incidence of morbidity in affected children [3]. Research into the pathogenic mechanisms of S. aureus in PSA is important to improve early diagnosis and effective treatment.
Objectives: To analyze the molecular and cellular mechanisms involved in pediatric septic arthritis caused by S. aureus, using transcriptomic data and immune cell profiling.
Methods: Dataset GSE30119 was obtained from the Gene Expression Omnibus platform, including transcriptional profiles of patients with community-acquired S. aureus infection. 19 patients with PSA and 44 healthy controls were included in the analysis. Differentially expressed genes (DEGs) were filtered using a false-discovery rate of <0.05 and a log2foldchange >1. Functional enrichment was performed using the Metascape portal. PPIs were studied on the Cytoscape platform, and hub genes (HGs) were determined using five algorithms with the Cytohubba plugin. MFI values were obtained for each patient and receiver operator characteristics (ROC) curves were drawn. The online tool CibersortX was used to predict immune cell types and abundance in peripheral blood. The proportion of immune cells between groups was determined using a two-way ANOVA test, and Sidak’s correction for multiple comparisons. A spearman test was conducted to correlate immune cells with HGs. A p value of < 0.05 was considered statistically significant.
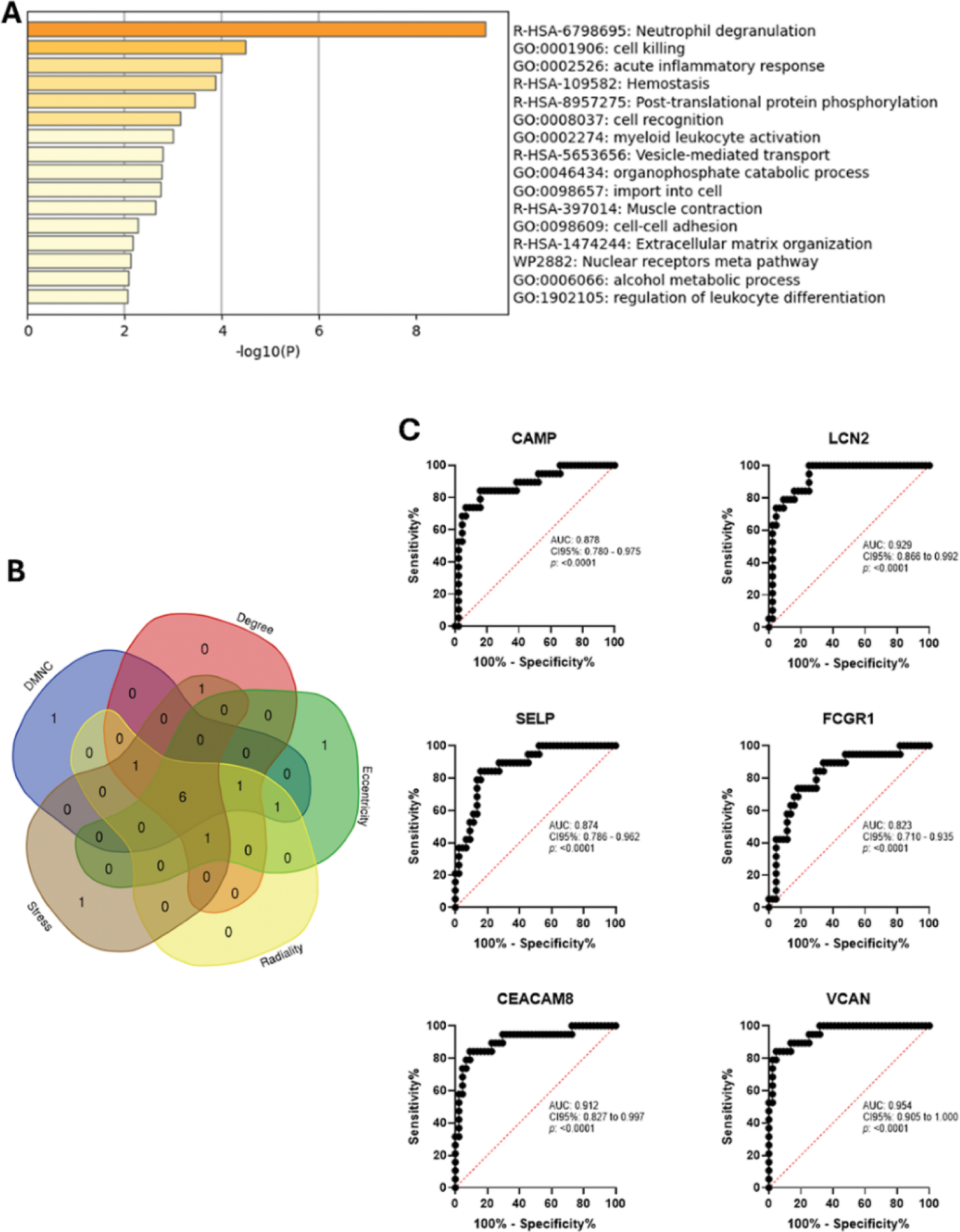
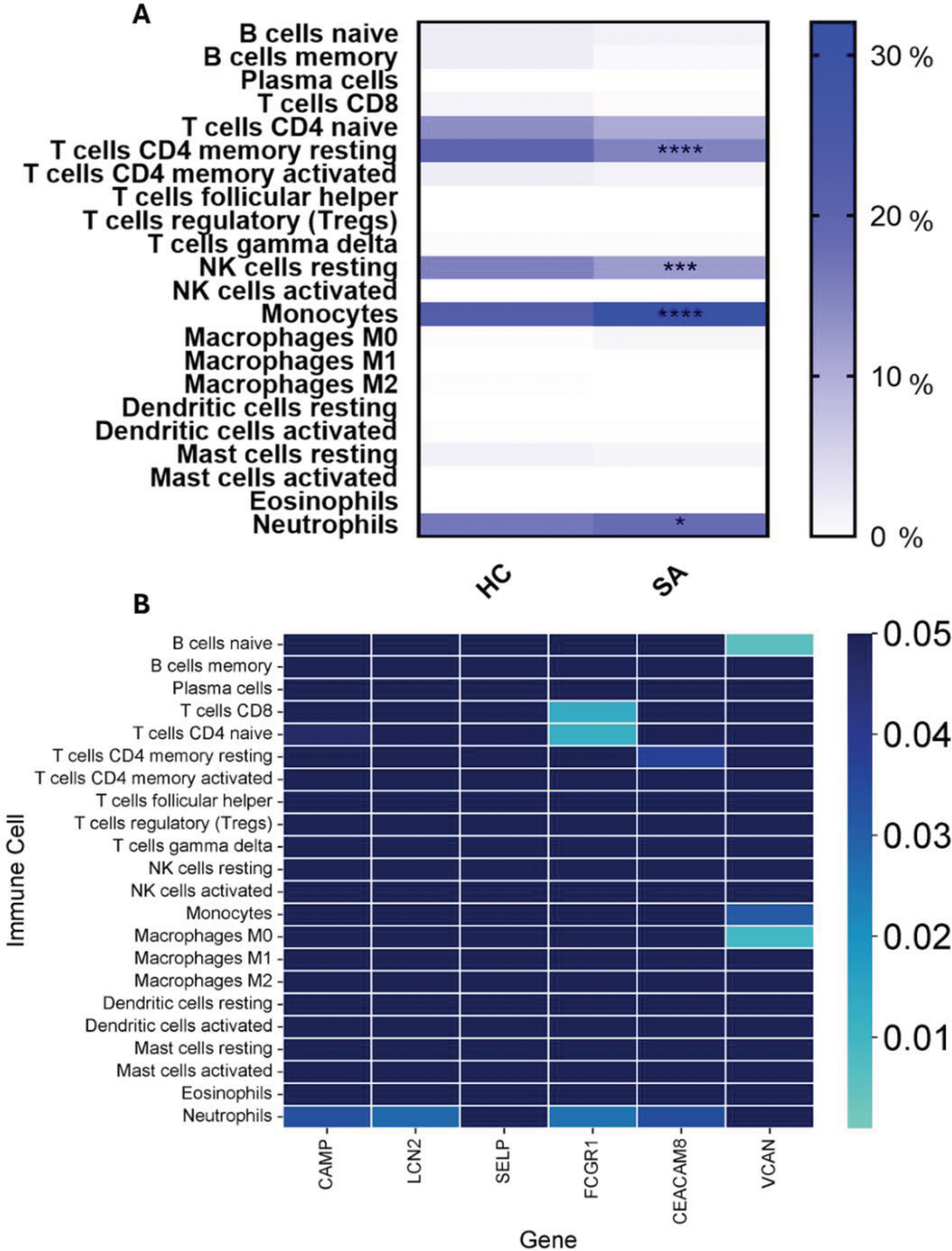
Results: 39 DEGs were identified. Functional enrichment analysis is shown in Figure 1A. CAMP , LCN2 , SELP , FCGR1 , CEACAM8 , and VCAN were identified as HGs. ROC curves presented appropriate areas under the curve for prompt discrimination between patients and controls. Significant differences in the levels of immune cell populations were found in the proportions of CD4 Memory Resting T Cells, Resting NK Cells, Monocytes, and Neutrophils. Significant associations were noted: CAMP and CD4 Naïve T Cells (r = -0.437) and neutrophils (r = 0.502); LCN2 and neutrophils (r = 0.517); FCGR1 and CD8 T Cells, (r = -0.572) and also with CD4 Naïve T Cells, (r = -0.576), and neutrophils,(r = 0.523); CEACAM8 and neutrophils, (r = 0.500), and also with CD4 Memory Resting T Cells, (r = -0.492); VCAN and Naïve B Cells, (r = -0.617), with Monocytes, (r = 0.508) and with M0 Macrophages, (r = 0.591).
Functional and Diagnostic Analysis of Key Genes in Pediatric Septic Arthritis. (A) Functional enrichment analysis of the 39 DEGs identified in PSA. The analysis revealed key biological processes, including neutrophil degranulation, cell killing, and acute inflammatory response. (B) Venn diagram illustrating the overlapping HGs through the five different algorithms (DMNC, Degree, Radiality, Eccentricity, Stress) in PSA. The overlapping genes ( CAMP , LCN2 , SELP , FCGR1 , CEACAM8 , and VCAN ) represent key targets involved in PSA pathogenesis. (C) Receiver Operating Characteristic (ROC) curves for the identified hub genes demonstrated diagnostic potential in differentiating PSA patients from healthy controls. Each ROC curve shows the area under the curve (AUC), providing a measure of the accuracy of these genes as potential diagnostic biomarkers.
Immune cell profiling based on LM22 . (A) Comparison of immune cell proportions between PSA and HC groups. Statistically significant differences are indicated by asterisks, where not shown p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (B) Heatmap displaying Spearman’s correlation analysis between HGs and immune cell populations. The color gradient indicates the significance of the correlation, where stronger blue indicates p > 0.05.
Conclusion: This study used transcriptomic profiles to analyze the pathogenesis of PSA driven by S. aureus. Several HGs with excellent diagnostic biomarker potential were identified, along with their relationship with immune cell populations. Neutrophils and other immune cells have been highlighted to play an imperative part in the pathogenesis of PSA. This will better justify the research to compare the transcriptomic profiles of patients infected with different causative microorganisms with a view to understanding the disease of PSA and improving targeted therapeutic strategies.
REFERENCES: [1] Mathews, C. J., Weston, V. C., Jones, A., Field, M., & Coakley, G. (2010). Bacterial septic arthritis in adults. The Lancet , 375(9717), 846-855.
[2] Goldenberg, D. L. (2017). Septic arthritis. The Lancet , 393(10180), 461-473.
[3] Salgado, C., Gomez, C. E., & Muldoon, K. (2020). Staphylococcus aureus infections in children: Pathogenesis, clinical manifestations, and treatment. Current Pediatric Reviews , 16(2), 100-109.
Acknowledgements: We’d like to thank the Mexican National Council of Humanities, Science and Technology (CONAHCYT) for providing the academic grant that made this research possible. Grant ID: PCC-2022-320697.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (