

Background: Rheumatoid arthritis (RA) is a long-term autoimmune condition with a complicated and varied underlying cause. The onset of RA has been demonstrated to be intimately linked to the Intestinal flora, which is crucial for immune regulation. Therefore, regulating the Intestinal flora provides an innovative alternative therapeutic strategy for alleviating RA symptoms.
Objectives: The purpose of this study is to further explore the relationship between RA and gut microbiota, find possible key flora and targets, and provide new evidence for microbiota therapy.
Methods: We performed 16srRNA sequencing on the feces of 38 new-onset RA patients and 22 healthy controls, assessed the microbiota diversity using Alpha and Beta diversity, and performed functional enrichment analysis using tools such as PICRUSt. Mendelian randomization (MR) was employed to examine the causal relationship between RA and alterations in the Intestinal flora.
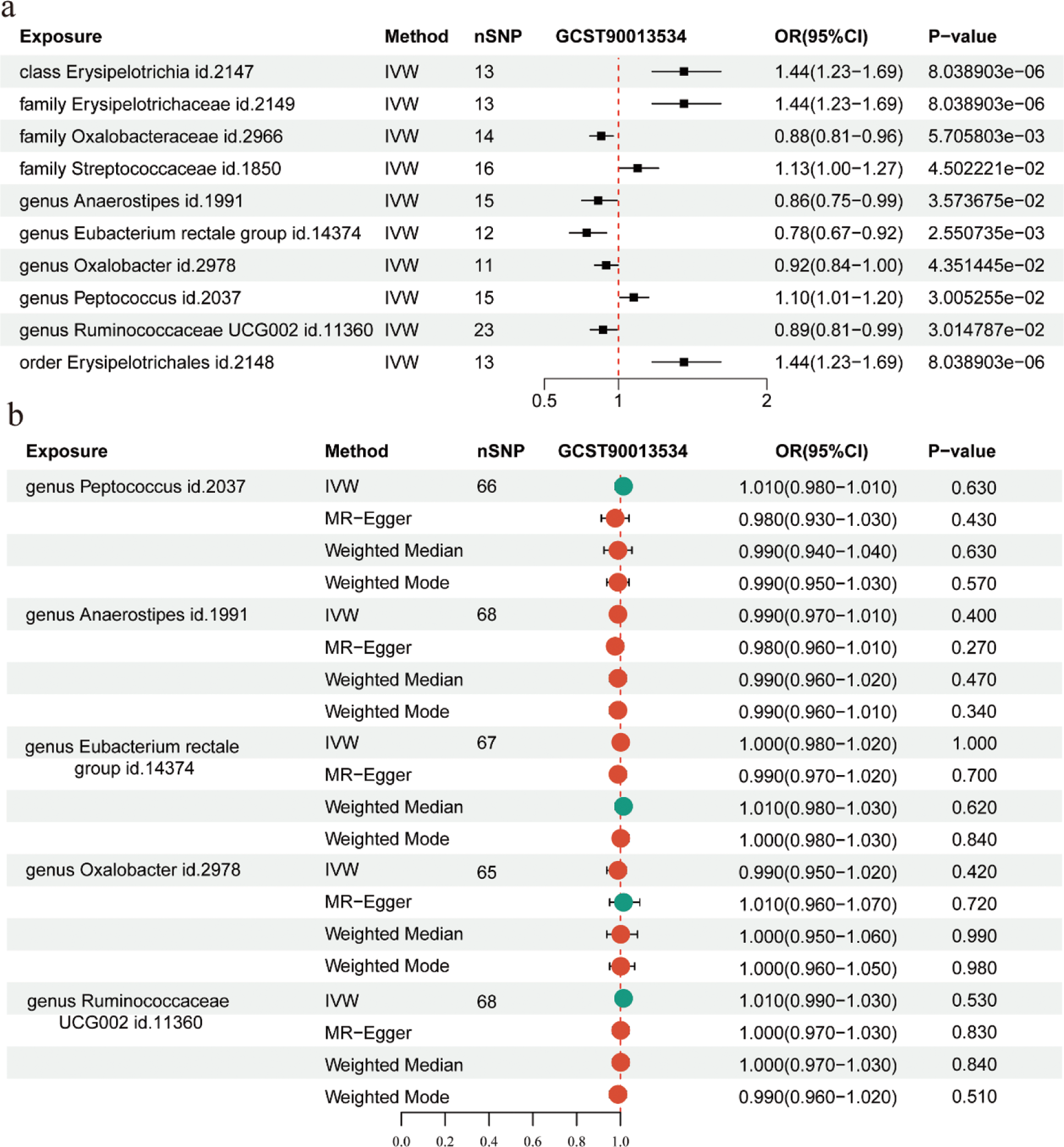
Results: A significant difference in the beta diversity of Intestinal flora was observed between RA patients and healthy controls(HC). Compared to the HC group, the genus level of Acidaminococcus and Gemella was upregulated in the gut of RA patients. Compared with the HC group, Bacteroidetes was down-regulated in RA at the phylum level, and Parabacteroides , Enterococcus , Butyricimonas , Oxalobacter , Christensenellaceae_R_7_group , Clostridium_sensu_stricto_1, Desulfovibrio , Actinomyces , Eubacterium_rectale_group , Erysipelotrichaceae_UCG−003 , Streptococcus , Lachnospiraceae_UCG−004 , Ruminococcaceae_UCG−004 , Peptococcus and Staphylococcus were down-regulated in RA at the genus level. The intestinal microbiota functions of RA patients were found to be enriched in pathways like genetic information processing and material metabolism, according to subsequent functional enrichment analysis. This suggests that the differential microbiota may have an immunomodulatory role through epigenetic and material metabolism signal transduction. Further random forest model was used to find that Firmicutes, Bacteroidetes, Actinobacteria, Lachnospiraceae_UCG.005, Roseburia, Pseudobutyrivibrio, Bifidobacterium, Faecalibacterium, Dialister, Ruminococcus_1, Eubacterium_rectale_group, Bacteroides were the genera with characteristic significance in RA patients. Anaerostipes and Eubacterium_rectale_group had a protective impact on RA, according to further MR research, but Erysipelotrichia and Streptococcaceae were associated with an elevated risk of RA. A causal relationship between these groups and RA was not shown by reverse MR analysis.
Conclusion: The study found that Firmicutes, Bacteroidetes, Actinobacteria, Eubacterium rectale group and other genera were associated with RA, especially Eubacterium_rectale_group , which showed significant significance in RA intestinal microbial flora and MR research. For further study on the pathogenesis of RA and the creation of novel therapeutic alternatives, these results offer crucial references.
Flowchart illustrating the main hypotheses and methods of the current study. And comparison of differential intestinal microbiota between new-onset rheumatoid arthritis (RA) patients and healthy controls and functional enrichment of intestinal microbiota. Differences were considered statistically significant at P < 0.05. HC group: healthy control group; PA group: new-onset rheumatoid arthritis patient group.
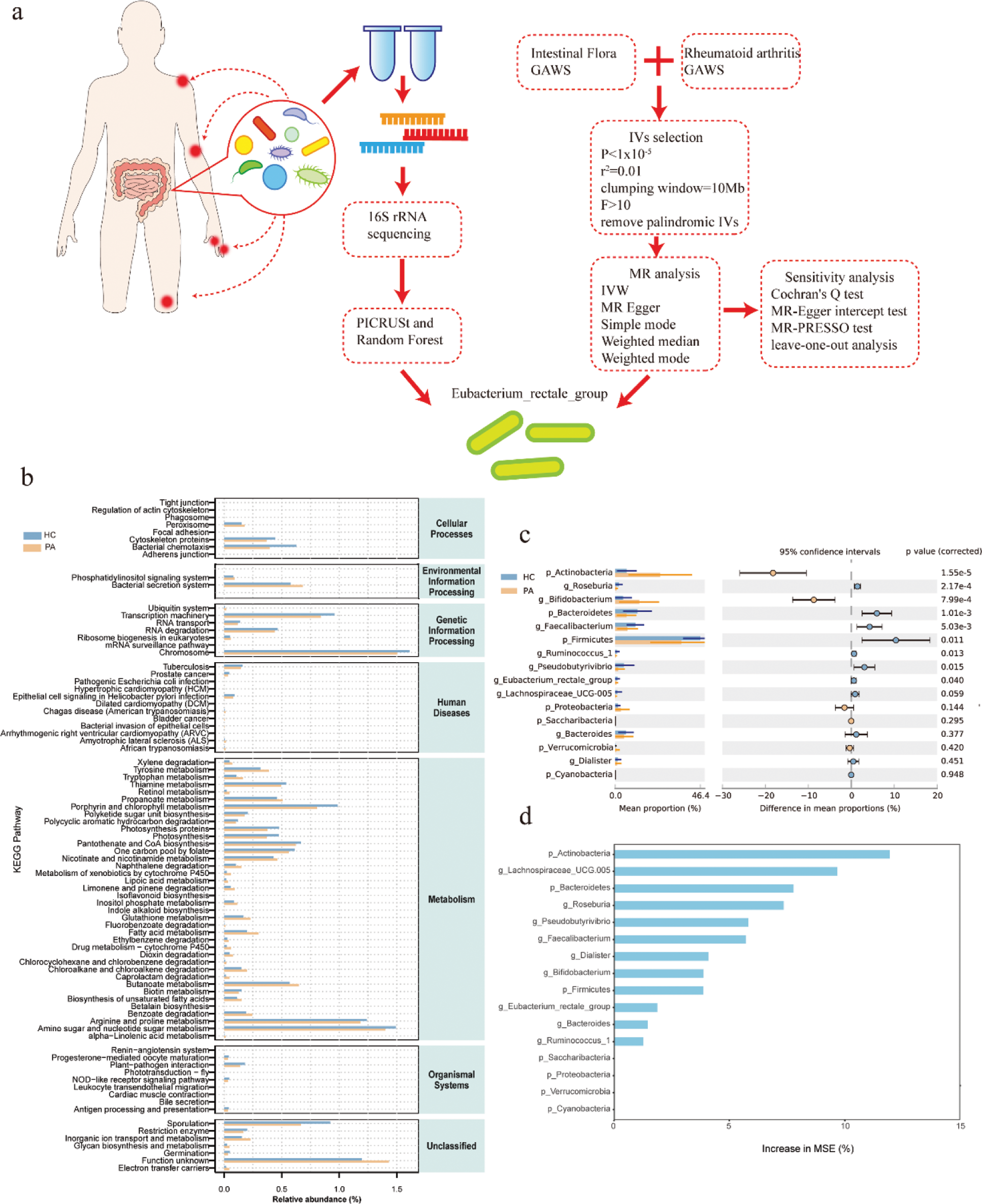
Mendelian randomization (MR) analysis of the causal relationship and the reverse causal relationship between Intestinal flora and rheumatoid arthritis (RA).
REFERENCES: [1] Lin YJ, Anzaghe M, Schülke S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells. 2020;9(4). Epub 2020/04/09. doi: 10.3390/cells9040880. PubMed PMID: 32260219; PubMed Central PMCID: PMCPMC7226834.
[2] Scherer HU, Häupl T, Burmester GR. The etiology of rheumatoid arthritis. Journal of autoimmunity. 2020;110:102400. Epub 2020/01/26. doi: 10.1016/j.jaut.2019.102400. PubMed PMID: 31980337.
[3] Di Matteo A, Bathon JM, Emery P. Rheumatoid arthritis. Lancet (London, England). 2023;402(10416):2019-33. Epub 2024/01/19. doi: 10.1016/s0140-6736(23)01525-8. PubMed PMID: 38240831.
[4] Gravallese EM, Firestein GS. Rheumatoid Arthritis - Common Origins, Divergent Mechanisms. The New England journal of medicine. 2023;388(6):529-42. Epub 2023/02/14. doi: 10.1056/NEJMra2103726. PubMed PMID: 36780677.
[5] Zaiss MM, Joyce Wu HJ, Mauro D, Schett G, Ciccia F. The gut-joint axis in rheumatoid arthritis. Nature reviews Rheumatology. 2021;17(4):224-37. Epub 2021/03/07. doi: 10.1038/s41584-021-00585-3. PubMed PMID: 33674813.
Acknowledgements: The authors are grateful to MiBioGen consortium, UK Biobank, GIANT consortium, GWAS Catalog, and IEU OpenGWAS project for their selfless public sharing of GWAS summary data, which provides us with great convenience to carry out this research.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (