

Background: Idiopathic inflammatory myopathy (IIM) is a rare systemic autoimmune disease with heterogenous phenotypes. While muscle biopsy is used to differentiate IIM phenotypes based on specific histological features, the underlying mechanisms remain poorly understood. Transcriptomic studies have provided valuable insights into IIM pathology, including the distinct upregulation of type I interferon (IFN)-inducible genes, notably in dermatomyositis (DM) and, to a lesser extent, in anti-synthetase syndrome (ASyS) [1]. Despite these advances, most studies rely on bulk RNA sequencing, with limited exploration using single-cell RNA sequencing, leaving gaps in understanding cellular-level heterogeneity.
Objectives: To explore the landscape of muscle-infiltrating immune cells in a multimodal scale at the single cell level in IIM patients who underwent muscle biopsy.
Methods: Muscle tissues from 18 IIM patients were obtained and CD45 positive cells were sorted for further single cell analyses. Cells were subjected to scRNA-seq, scCITE-seq, scTCR-seq and scBCR-seq using the 10x Genomics Chromium platform. 10 tested positive for anti-aminoacyl t-RNA synthetase (ARS) antibody, four tested positive for anti-Mi2 antibody, one tested positive for anti-MDA5 antibody, and one tested positive for anti-transcriptional intermediary factor 1-γ (TIF1-γ). Two had no myositis specific antibodies (MSA). Unsupervised clustering was performed based on both RNA-seq data and CITE-seq data, using weighted nearest neighbor method with graph-based clustering and uniform manifold approximation and projection (UMAP) visualization. Sub-clustering was performed on CD4+ T cells and CD8+ T cells. Clusters were annotated manually by the marker genes of known subtypes of immune cells. Associations between each cluster and clinical features or pathological features of muscle biopsy were examined by generalized linear mixed model (GLMM). Cell-cell communication was analyzed with CellChat. Additionally, TCR and BCR repertoire analyses were performed.
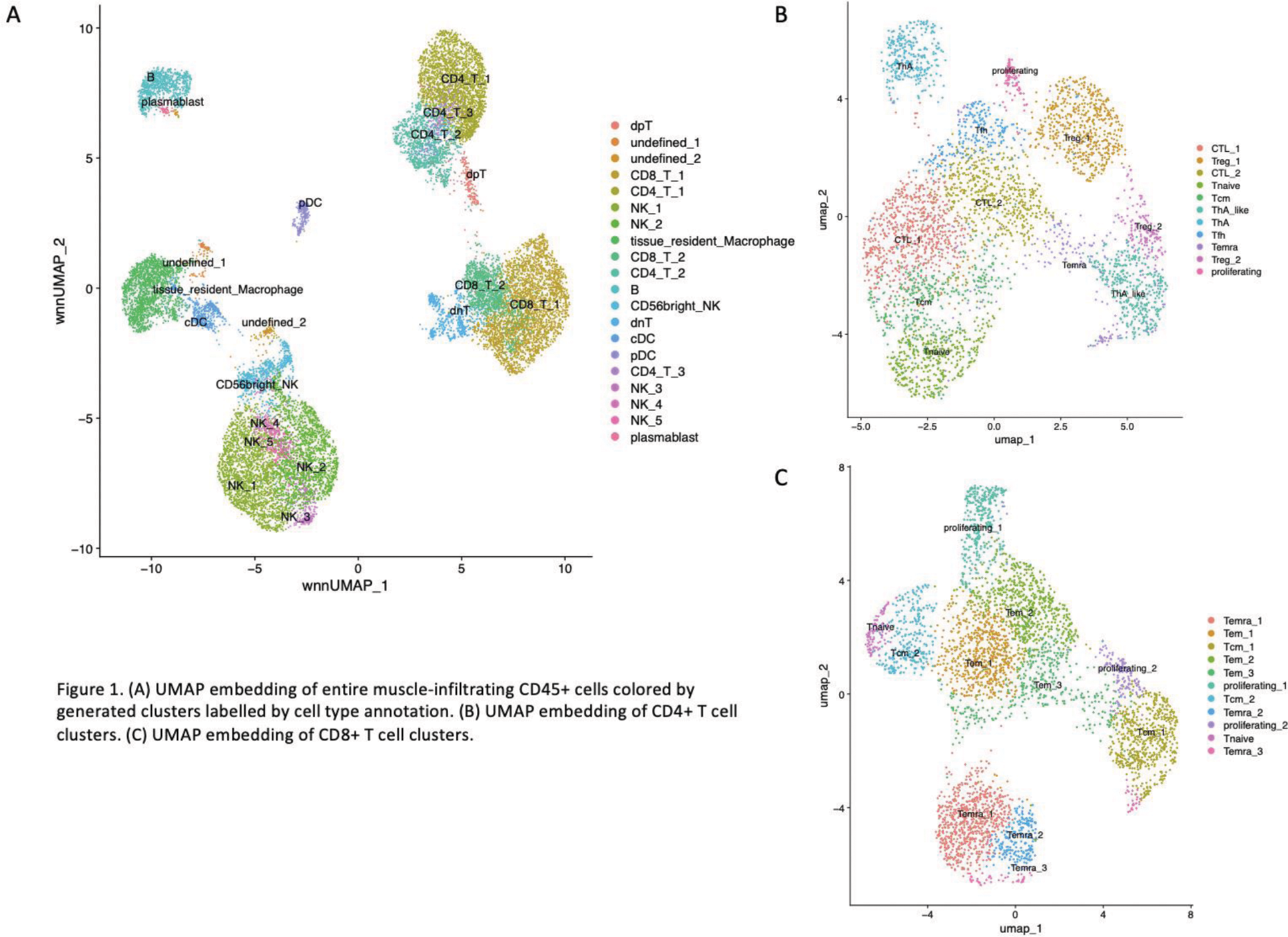
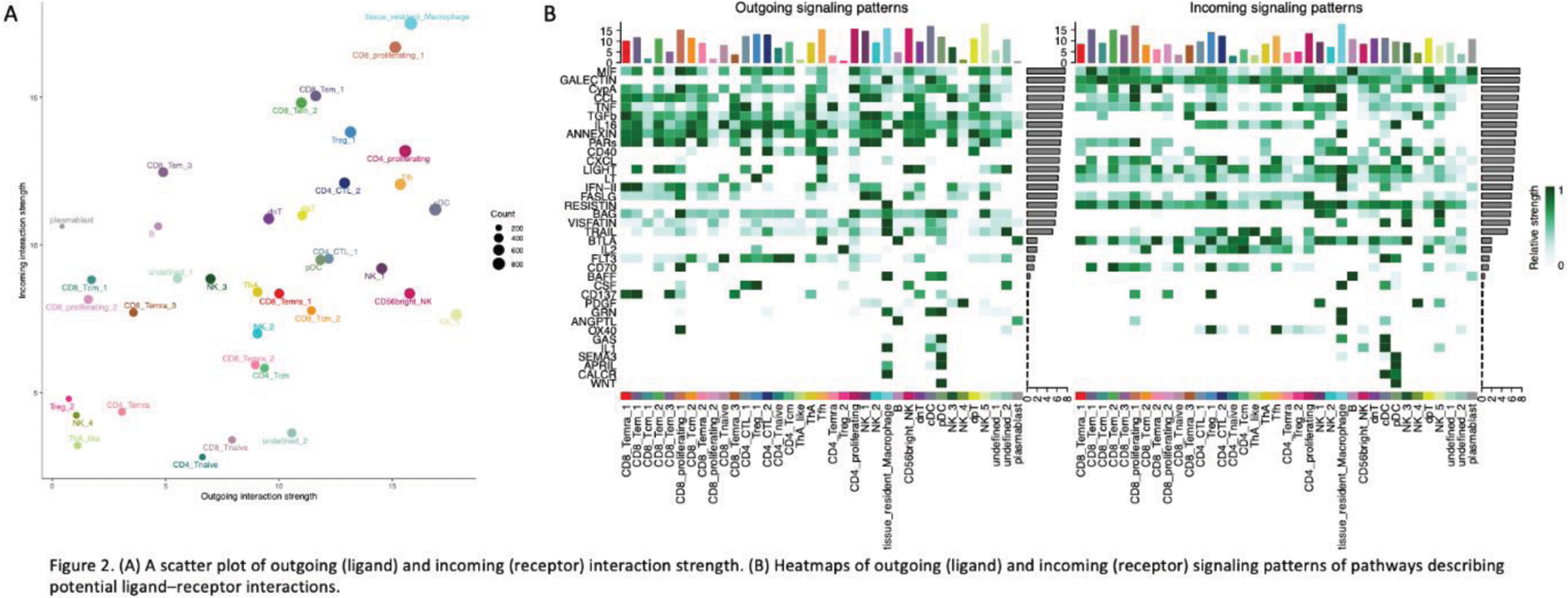
Results: A total of 20 immune cell clusters were identified from muscle-infiltrating immune cells (Figure 1A). Sub-clustering of CD4+ T cells revealed the presence of age-associated helper T cells (ThA) (Figure 1B), a population known for both cytotoxic and B cell help functions [2]. ThA was the most clonally expanded cluster among CD4+ T cell subsets. Sub-clustering of CD8+ T cells identified 10 sub-clusters (Figure 1C), with the terminally differentiated effector memory T cell cluster 1 (CD8_Temra_1) showing prominent clonal expansion. In contrast, most cells in the B cell cluster exhibited a single clone. Cell-cell communication analysis revealed tissue-resident macrophages as the cluster with the strongest incoming interactions (Figure 2A), particularly characterized by the incoming IFNG (IFN-II) signal (Figure 2B). In terms of outgoing interactions, NK_5 exhibited the strongest activity (Figure 2A). Among CXC motif chemokines, the CXCL13-CXCR3 pair was the dominant interaction, with CXCL13 primarily expressed in the follicular helper T cell (Tfh) cluster. The CXCL10-CXCR3 pair was the second most prominent, with CXCL10 predominantly secreted by tissue-resident macrophages, likely stimulated by interferon-gamma (IFN-γ). Generalized linear mixed model (GLMM) analysis demonstrated a positive correlation between anti-ARS antibody positivity and the CD8_Temra_1, CD8_Temra_3, and CD8+ proliferating T cell clusters (CD8_T_proliferating_1) (p=0.038, p=0.035, and p=0.034, respectively). In contrast, anti-Mi-2 antibody positivity was negatively correlated with CD8_Temra_1, CD8_Temra_2, CD8_Temra_3, and ThA (p=0.0003, p=0.009, p=0.0008, and p=0.002, respectively). Regarding muscle pathological features, complement complex deposition in small intramuscular sheath vessels, a hallmark of dermatomyositis (DM), was positively correlated with NK_4 and NK_5 clusters (p=0.042 and p=0.008, respectively). Perifascicular atrophy, another characteristic DM feature, was positively correlated with NK_4 and NK_5 (p=0.024 and p=0.011) and negatively correlated with CD8_Temra_1, CD8_Temra_2, CD8_Temra_3, and ThA (p=0.021, p=0.020, p=0.018, and p=0.013, respectively).
Conclusion: Single-cell transcriptomics combined with repertoire analysis of muscle-infiltrating immune cells revealed distinct alterations in immune cell subtype abundance, particularly in ASyS and DM. Clonally expanded CD8+ Temra cells emerged as a characteristic cluster in ASyS but were less prevalent in Mi-2-positive DM. Additionally, pathological features associated with DM showed positive correlations with specific NK cell clusters. These findings advance our understanding of the immunopathogenesis of IIM and lay the groundwork for further exploration of muscle-infiltrating immune cells in IIM.
REFERENCES: [1] Pinal- Fernandez I, Casal-Dominguez M, Derfoul A, et al. Identification of distinctive interferon gene signatures in different types of myositis. Neurology. 2019;93:e1193–e1204.
[2] Goto M, Takahashi H, Yoshida R, et al. Age-associated CD4+ T cells with B cell-promoting functions are regulated by ZEB2 in autoimmunity. Sci Immunol. 2024;9:eadk1643.
Acknowledgements: This research was supported by the Japan Agency for Medical Research and Development (AMED).
Disclosure of Interests: Tomoya Yoshida: None declared , Hideyuki Takahashi: None declared , Takahiro Itamiya Chugai, Takeshi Iwasaki: None declared , Natsuka Umezawa Abbvie Japan Co., Ltd., Astellas Pharma Inc., Boehringer Ingelheim, Taisho Pharmaceutical Co., Ltd., Kyowa Kirin Co., Ltd., Hirokazu Sasaki Abbvie, Asahi Kasei Pharma, and Mitsubishi Tanabe Pharma, Taisho Pharmaceutical Co., Ltd and Kissei Pharmaceutical Co., Ltd., Keita Ninagawa: None declared , Yuichiro Fujieda MEDICAL & BIOLOGICAL LABORATORIES CO., LTD., Yusuke Miyazaki Eli Lilly, GlaxoSmithKline, AstraZeneca, Astellas, Asahi-kasei, UCB, Yasuyuki Kamata: None declared , Masaru Takeshita: None declared , Masakazu Matsushita: None declared , Toshio Kawamoto: None declared , Meiko Maeda: None declared , Akatsuki Kubota: None declared , Tatsushi Toda: None declared , Naoto Tamura ASAHI KASEI PHARMA Corp., AstraZeneca plc, AbbVie Inc., Eli Lilly Japan K.K, GlaxoSmithKline K.K., Novartis Pharma K.K., Novartis Pharma K.K., Bristol Myers Squibb Co., Ltd, UCB Japan Co. Ltd, ASAHI KASEI PHARMA Corp., Ayumi Pharmaceutical Corp., AbbVie Inc., Taisho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Nippon Boehringer Ingelheim Co., Ltd., Yuko Kaneko: None declared , Tsutomu Takeuchi: None declared , Kojiro Sato Ono Pharma, GlaxoSmithKline, Chugai, Asahi Kasei Pharma, Shingo Nakayamada GlaxoSmithKline, Astellas Pharma, Eli Lilly, AbbVie, Mitsubishi Tanabe Pharma, Janssen, Pfizer, Bristol Myers Squibb, Asahi-Kasei, Chugai Pharmaceutical, AstraZeneca, Taisho Pharmaceutical, Eisai, Gilead Sciences, UCB, and Ayumi, Asahi-Kasei, Otsuka Pharmaceutical and Mitsubishi Tanabe Pharma, Yoshiya Tanaka AbbVie, Eisai, Chugai, Eli-Lilly, Behringer-Ingelheim, GlaxoSmithKline, Taisho, AstraZeneca, Daiichi-Sankyo, Gilead, Pfizer, UCB, Asahi-kasei, Astellas, Behringer-Ingelheim, Taisho, Chugai, Ken Yoshida AbbVie Inc., Asahi Kasei Pharma Corporation, Astellas Pharma Inc., Eisai Co., Ltd., Boehringer Ingelheim, Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Japan Blood Products Organization, Otsuka Pharmaceutical Co., Ltd., Taisho Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd, Tatsuya Atsumi Chugai, Astellas, Takeda, Mitsubishi Tanabe, Chugai, Pfizer, Eisai, AbbVie., Takeda, Mitsubishi Tanabe, Chugai, Pfizer, Daiichi Sankyo, Otsuka., Shinsuke Yasuda Eisai, ImmunoForge, Novartis, and Otsuka seiyaku, Asahi Kasei Pharma, Nippon Boehringer Ingelheim Co., Ltd, and Taisho Pharmaceutical Co., Ltd, speaking fees from Abbvie, Asahi Kasei Pharma, Chugai Pharmaceutical, Eisai, Eli Lilly, GlaxoSmithKline, Mitsubishi Tanabe Pharma, and Ono pharmaceutical, Koichiro Ohmura Otsuka Pharmaceutical Co., Ltd., Taisho Pharmaceutical Co., Ltd., AbbVie Japan GK, Asahi KASEI Pharma Co., Ltd., Astellas Pharma Inc., AstraZeneca plc., Ayumi, Boehringer Ingelheim Japan, Inc., Bristol Myers Squibb Co., Ltd., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Eli Lilly and Company, Gilead Sciences, Inc., GlaxoSmithKline K.K., Mitsubishi-Tanabe Pharma Co., Ltd., Novartis Pharma K.K., Otsuka Pharmaceutical Co., Ltd., Janssen Pharmaceutical K.K., UCB Japan Co. Ltd., AbbVie Japan GK, Asahi KASEI Pharma Co., Ltd., Chugai Pharmaceutical Co., Ltd., Mitsubishi-Tanabe Pharma Co., Ltd., Akio Morinobu Abbvie, Asahi Kasei, Astellas, Brystol Meyers, Chugai, Eisai, Eli Lilly, Tanabe Mitsubishi, Pfizer, Kazuhiko Yamamoto Chugai Pharmaceutical CO., LTD., Tomohisa Okamura Asahi Kasei, EpiVax, Bristol-myers Squibb, Chugai, Keishi Fujio Chugai, Abbvie, Asahikasei Pharma, Bristol Myers, AstraZeneca, Tanabe Mitsubishi, Eisai, Gilead, Eli Lilly, Pfizer, Taisho, Astellas, Daiichi-Sankyo, Novartis, GlaxoSmithKline, Alexion Pharma, Asahikasei Pharma, Chugai, Asahikasei Pharma, AstraZeneca, Tsumura, Abbvie, Bristol Myers, Eisai, Taisho.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (