

Background: Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease. Diagnosis of RA relies on clinical and laboratory data while the search for more precise biomarkers in RA is ongoing to improve early identification and offer timely and effective personalized treatment. A genomic transformer is a novel (since 2023), robust and scalable artificial intelligence (AI) foundation model for genomics that leverages transformer architecture to use and interpret exomic data as text for improved prediction and analysis of DNA sequences. Only four genomic transformers have been published thus far. The most recent transformer, the Nucleotide Transformer, published in November 2024, is a decoder-only Generative Pretrained Transformer (GPT) style transformer, which outperformed existing models but similar to the previous models, has not been tested on clinical use cases in general and in rheumatology specifically.
Objectives: Leveraging the large biobank of exomic data, we aimed to 1) develop and validate a novel exomic transformer to process and interpret exomic information; and 2) apply the nucleotide transformer to identify sequences associated with RA from a cohort of research participants.
Methods: This project leverages the Mayo Clinic Cloud (MCC ) and the Cerebras Cluster for computational needs, required to process over 4 terabytes of exomic data. The Tapestry Cohort is a large, decentralized, clinical Exome+ assay study of 98,222 individuals [2] and is used as the primary source of exomic data for this study. NCBI human reference genome data and data on multispecies were also used for structural element training. NIH/NCBI-hosted ClinVar data on human genetic variants and their relevance to certain diseases were used for functional training. Within the Tapestry cohort we have identified a cohort of adult (aged >/=18 years) persons with RA (n=5,984) that was manually reviewed for confirmation of their RA diagnosis. To create the exomic transformer, data were prepared by converting Binary Alignment Map (BAM) files to Browser Extensible Data (BED) and FASTQ files, followed by filtration and compression to reduce file size without information loss. The tokenization process was used to convert nucleotide sequences into integer values for transformer processing. The transformer was constructed using an autoregressive, decoder-only architecture, pretrained on the next token prediction task. The model focused on unique subject’s genetic information within the context of the human reference genome. To optimize the model’s performance, multiple experiments were conducted with modification in model’s components and model size (i.e., ablation and scaling). Evaluation metrics included the Matthews Correlation Coefficient (MCC) for upstream model evaluations and structural component evaluation for downstream tasks. We used the clinical applications that were created for the Mayo exomic transformer to benchmark similar clinical phenotypes on the four other publicly available transformer models that had no prior clinical applications. The exomic transformer was trained on the NCBI human reference genome (HRG38). In addition 768 reference genomes from multiple human, animal and bacterial species were used for training to enable recognition of key genomic structures. Then a sample of 507 patients from the Tapestry cohort was chosen (160 with RA and 347 without RA) and their exome sequences were added to the training data.
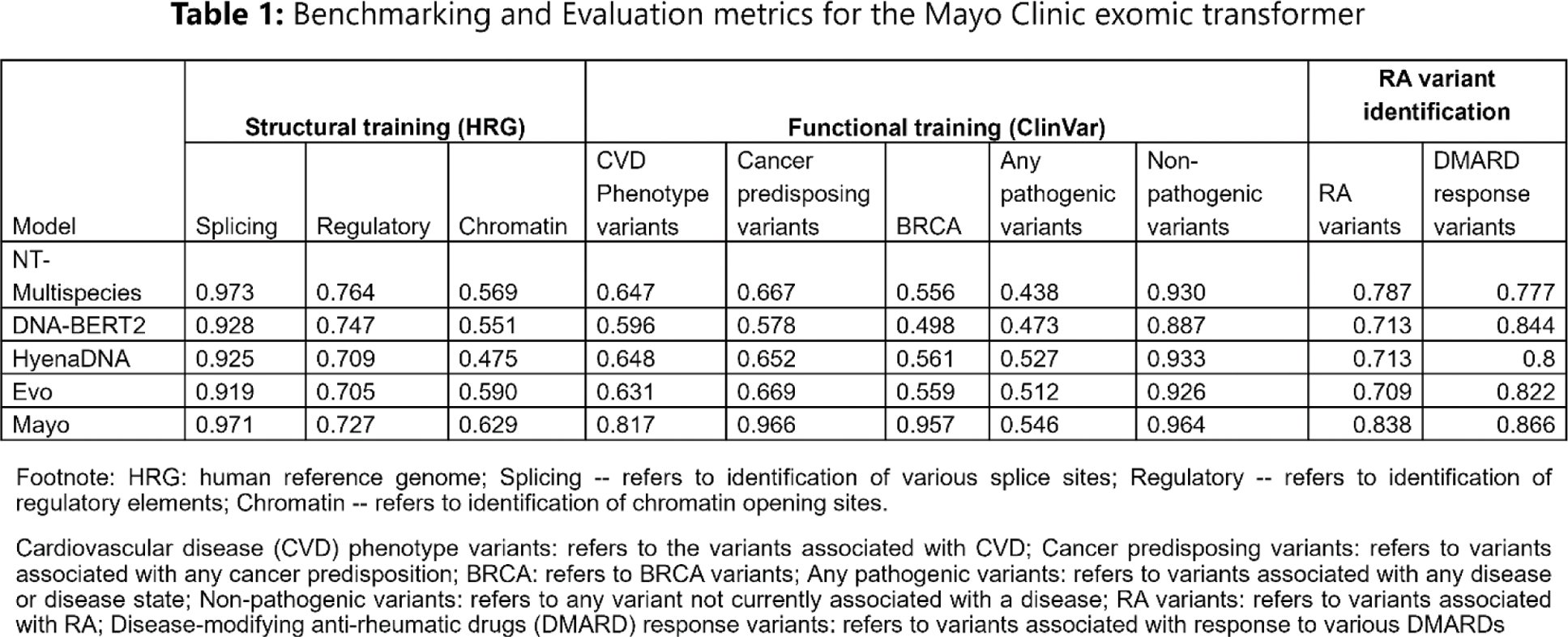
Results: The final model has 1 billion parameters and 20 layers with a context length of 1000 nucleotide bases. The model architecture follows LLaMA-2 model architecture, one of the most successful models for text data. The overall training dataset contained more than 1 trillion tokens, with one token representing a single nucleotide base. The exomic transformer achieved excellent accuracy in identifying structural genetic elements (0.971 for splicing sites, 0.727 for regulatory sites). Testing with functional elements from ClinVar showed excellent performance for identifying the following phenotypes: 0.817 for cardiovascular phenotyping, 0.966 for cancer syndromes. A set of RA specific evaluations was then performed where the model was given sequences containing over 600 variants known to be associated with RA or response to treatment with antirheumatic drugs, and 2200 variants not associated with RA. From these variants the model’s accuracy was 0.791 in identifying RA specific variants and 0.866 for variants associated with response to various disease-modifying antirheumatic drugs. Table 1 summarizes the performance metrics of the Mayo Clinic exomic transformer compared to the metrics of the four existing transformers, including the benchmarked clinical applications that we created for these models. The results show that Mayo transformer has mostly superior performance across the structural and functional tasks.
Conclusion: We have developed and validated a first-in-class novel exomic transformer with a clinical application in RA. This transformer is able to process and accurately interpret structural and functional genetic variants. This study presents a proof of concept in utilizing a novel exomic transformer for processing exomic information, expanding the capabilities of AI use in medicine and rheumatology. The work on testing this transformer’s performance for identification of patients with RA among the entire Tapestry repository is currently underway and is envisioned to assist in early and efficient identification of persons with genetic make up for RA in large Biobanks. The applications of this AI tool are broad and studies on testing the transformer for the use in other chronic conditions and for prediction of antirheumatic medication response are ongoing.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (