

Background: Autoimmune disorders (ADs) are increasing in prevalence and leading to significant treatment costs. Emerging evidence from large-scale observational studies and meta-analyses suggests an increased risk of cardiometabolic diseases (CMDs) in patients with ADs. However, observational studies are subject to well-known biases, such as confounding due to unmeasured confounders and are prone to reverse causation. Mendelian randomisation (MR) mimics randomised controlled trials and provides a robust alternative for investigating potential causal relationships using genetic variants as instrumental variables.
Objectives: This study aimed to (1) elucidate the potential causal associations between ADs and CMDs, and (2) identify and characterise the plasma proteins important to the relationship between these two groups of diseases.
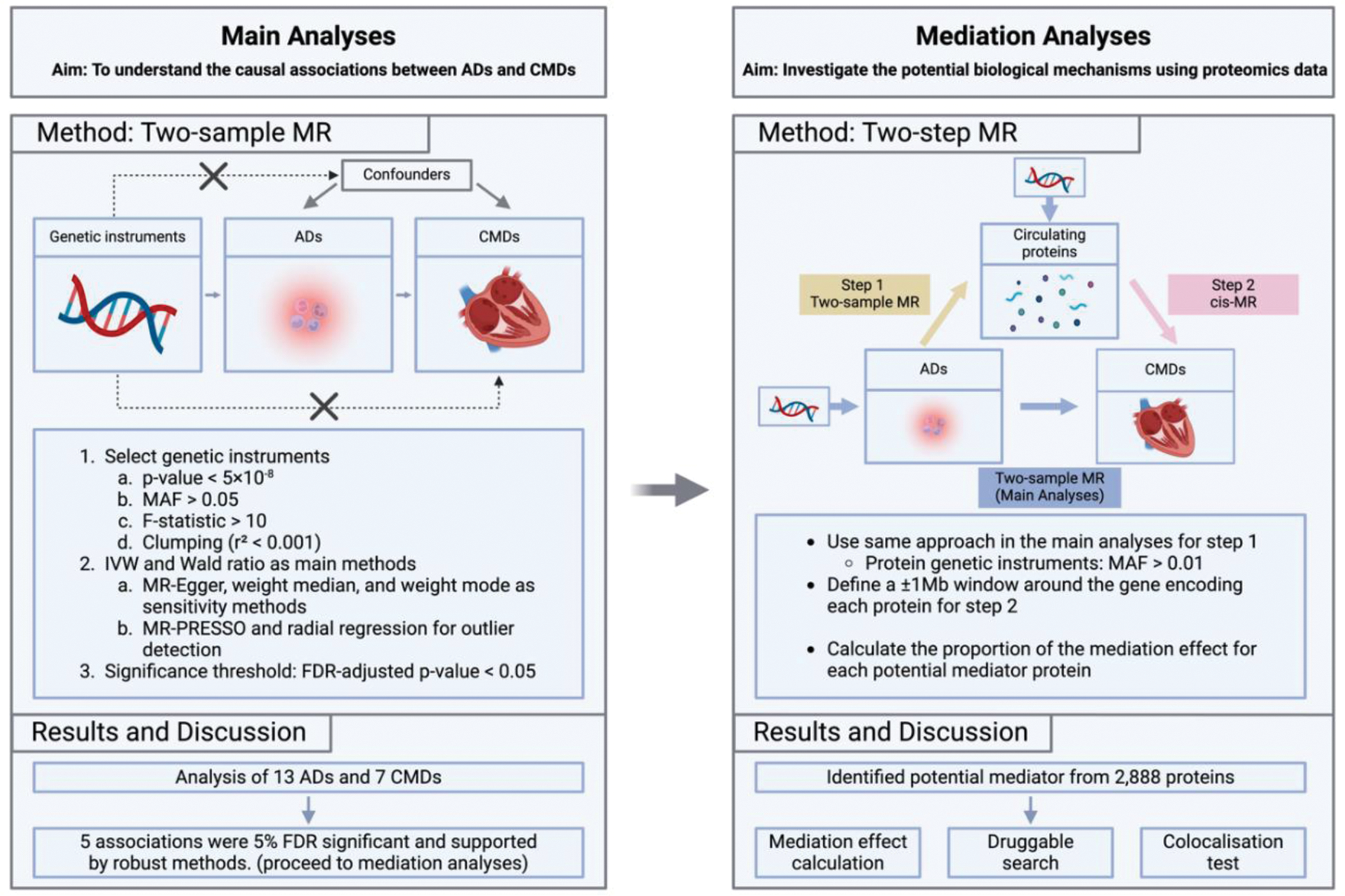
Methods: Figure 1 illustrates the study design, including the univariate MR and mediation analyses. The study utilised data from 21 genome-wide association studies (GWAS), covering 13 ADs, 7 CMDs, and one focused on proteins, with a total sample size exceeding 9 million individuals. Genetic associations for plasma proteins were obtained from the UK Biobank Pharma Proteomics Project, using an antibody-based Olink Explore 3072 proximity extension assay to measure around 2,900 proteins [1]. This study employed two-sample MR, using the Wald ratio and inverse variance weighted (IVW) methods as the primary approaches [2]. Robust methods, including MR-Egger, weighted median, and weighted mode, were also applied to account for potential pleiotropy [3, 4]. Furthermore, MR-PRESSO was applied to remove potential outliers, and single nucleotide polymorphisms (SNPs) in the human leukocyte antigens (HLA) region on chromosome 6 were excluded due to their complex linkage disequilibrium structure. Radial regression was also employed as an additional outlier detection method and to manage heterogeneity [5]. Subsequently, two-step MR was performed to identify proteins that act as potential mediators and utilised the colocalisation test with a posterior probability of H4 (PP.H4) > 80% as strong evidence and 60% < PP.H4 < 80% as potential evidence to support the results [6]. False discovery rate (FDR) correction was applied for multiple testing at 0.05 level.
Study design of two-sample MR for main analyses and two-step MR for mediation analyses. AD, autoimmune disorders; CMD, cardiometabolic diseases. Created with Biorender.com.
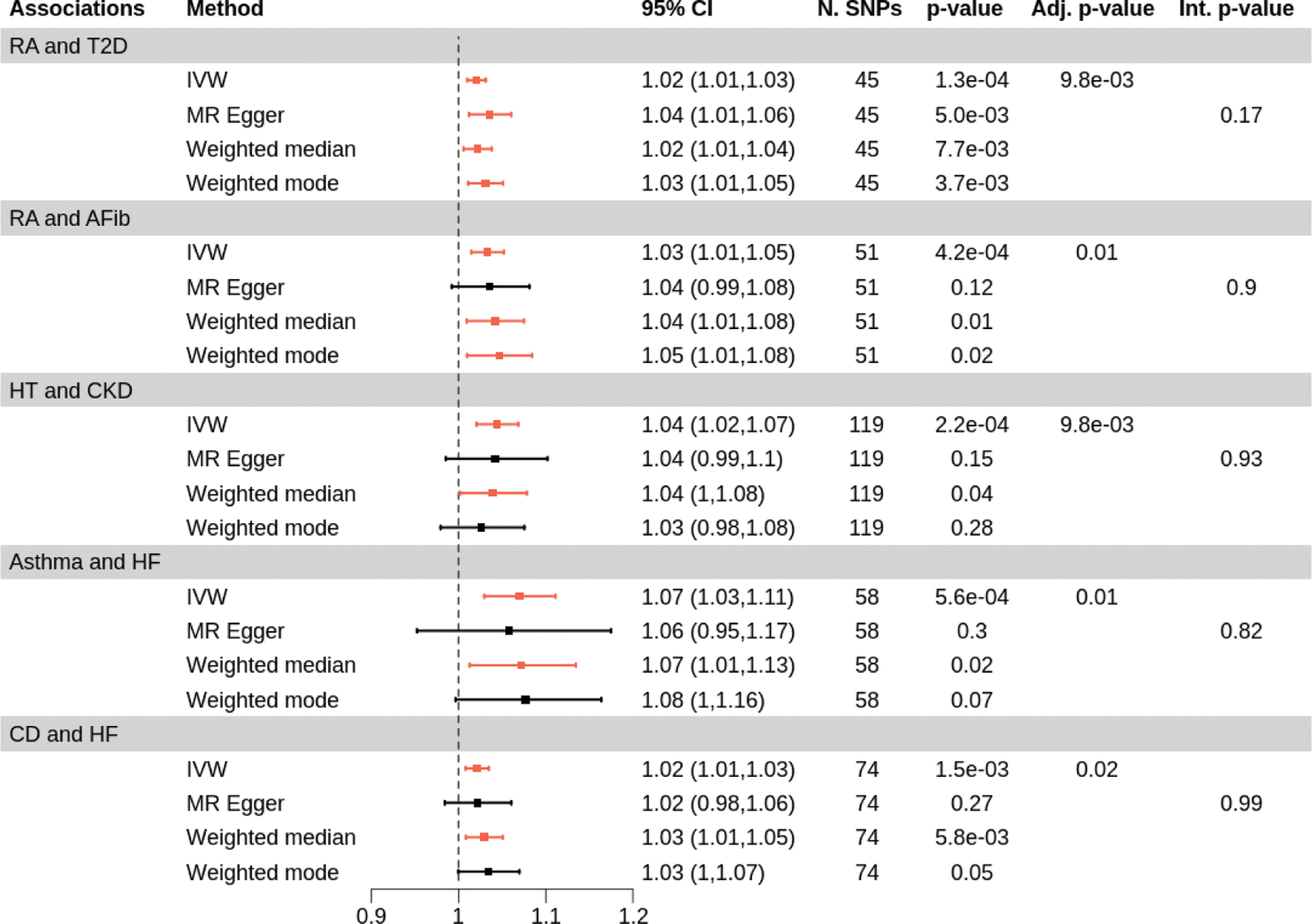
Results: This study investigated 91 associations between ADs and CMDs, of which 5 remained statistically significant after FDR correction. The odds ratios from IVW and Wald ratio estimates for these associations ranged from approximately 0.31 to 1.23, with the number of instruments varying from 1 to 130. Figure 2 demonstrates the details of five significant associations. Rheumatoid arthritis (RA) was linked to an increased risk of developing type 2 diabetes (T2D; OR IVW =1.02, 95% confidence interval [CI]: [1.01, 1.03]). The association was consistently supported by robust methods, including MR-Egger, weighted median, and weighted mode. A positive association was observed between RA and atrial fibrillation (OR IVW =1.03; 95%CI: [1.01, 1.05]), as well as between Hashimoto’s thyroiditis and chronic kidney disease (OR IVW =1.04; 95%CI: [1.02, 1.07]). Additionally, asthma (OR IVW =1.07; 95%CI: [1.03, 1.11]) and Crohn’s disease (OR IVW =1.02; 95%CI: [1.01, 1.03]) were both positively associated with heart failure. In any of these five associations, MR-Egger regression did not detect strong evidence of pleiotropy (MR-Egger intercept p -values > 0.05). In the mediation analysis, RBP1 and TNFRSF9 were potential mediators between RA and T2D, accounting for 37% (95%CI: [7%, 68%]) and 59% (95%CI: [14%, 104%]) of the mediation effect, respectively. This evidence was further supported by colocalisation with RBP1 (PP.H4 > 95%) and TNFRSF9 (PP.H4 > 77%).
Forest plots for 5 FDR significance associations. RA, rheumatoid arthritis; T2D, type 2 diabetes; AFib, atrial fibrillation; HT, Hashimoto’s thyroiditis; CKD, chronic kidney disease; HF, heart failure; Adj. P, adjusted p-value; N. SNPs, number of SNPs; Red, significant; Black, insignificant.
Conclusion: This study enhances our understanding of the potential causal associations between ADs and CMDs and the role of plasma proteins. Future research could explore these associations to identify drug targets, biological mechanisms, and preventive strategies for reducing cardiometabolic risks in autoimmune disease patients.
REFERENCES: [1] Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622 , 329-338, doi:10.1038/s41586-023-06592-6 (2023).
[2] Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37 , 658-665, doi:10.1002/gepi.21758 (2013).
[3] Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44 , 512-525, doi:10.1093/ije/dyv080 (2015).
[4] Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol 40 , 304-314, doi:10.1002/gepi.21965 (2016).
[5] Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51 , 957-972, doi:10.1038/s41588-019-0407-x (2019).
[6] Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J R Stat Soc Series B Stat Methodol 82 , 1273-1300, doi:10.1111/rssb.12388 (2020).
Acknowledgements: The authors thank the staff and participants of the UK Biobank, GWAS Catalog, FinnGen, IMSG, DIAGRAM, CARDIoGRAMplusC4D, and HERMES consortia for their efforts in generating and organising the invaluable data that were essential to this research.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (