

Background: Unpredictability is a major challenge in systemic lupus erythematosus (SLE). Routinely used clinical and laboratory parameters fail to predict the risk of and time to flare, or the type of flare that a patient might develop. We hypothesise that molecular biosignatures may better predict flaring and might thus have merit in disease monitoring.
Objectives: To investigate transcriptional biosignatures in relation to long-term flares in a well-characterised SLE population of European origin.
Methods: Eligible for this analysis were SLE patients from the European multicenter PRECISESADS project (NCT02890121) with available transcriptome data and long-term follow-up including registration of disease flares. The analysis was restricted to patients with quiescent disease at the time of sampling, defined as a clinical SLEDAI-2K score <6. Flare was defined as any increase in disease activity resulting in a change of therapy. Time-dependent analysis for interval censored data was conducted with semi-parametric models correcting for relevant confounding covariates associated with flares and individual weights according to limma/voom. Results were deemed significant if they yielded an FDR-corrected p-value <0.05 and a hazard ratio (HR) >1.5 for causative genes or <0.667 for protective genes. Gene ontologies were explored for genes yielding significant results using the ShiniGo web app.
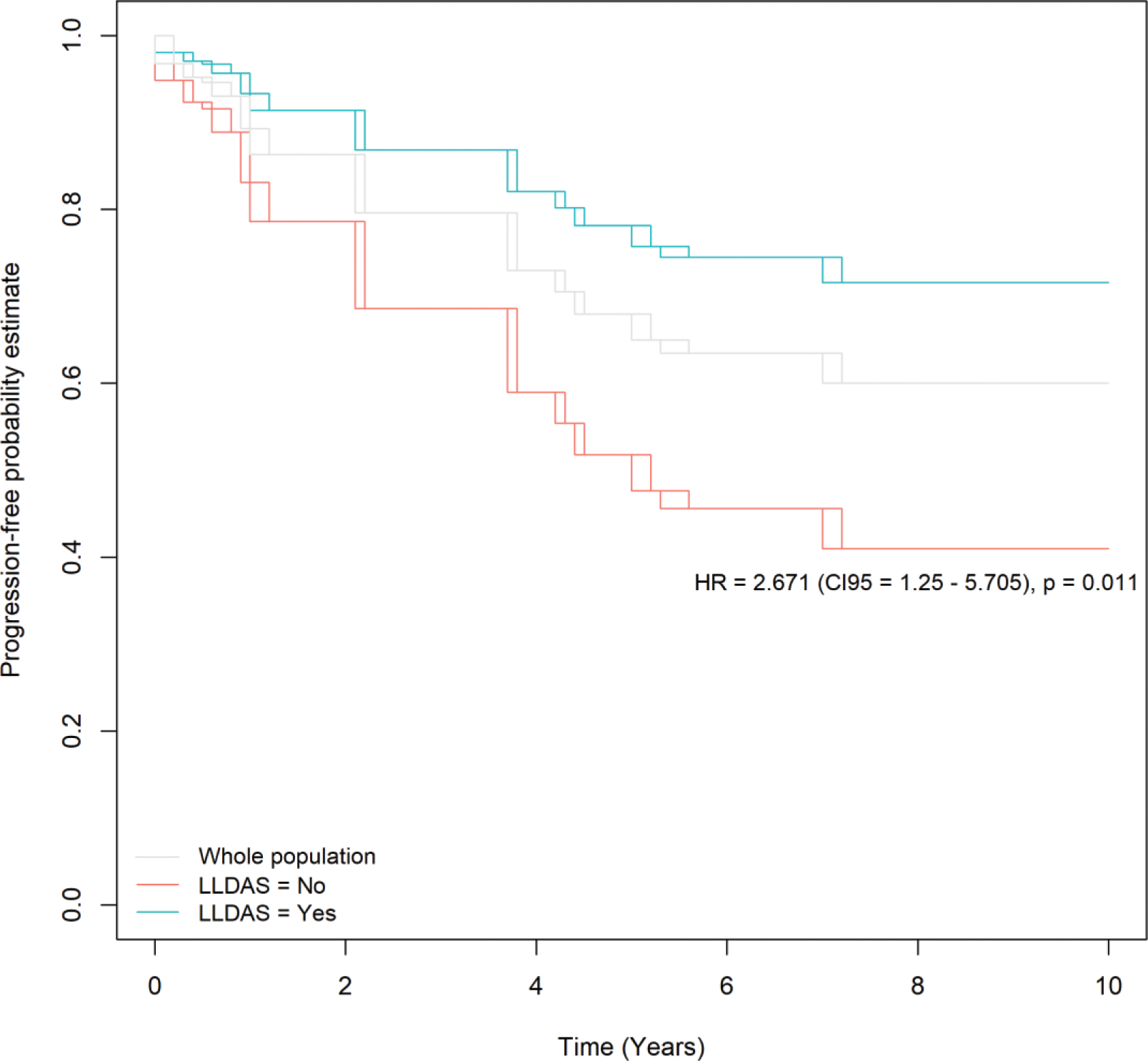
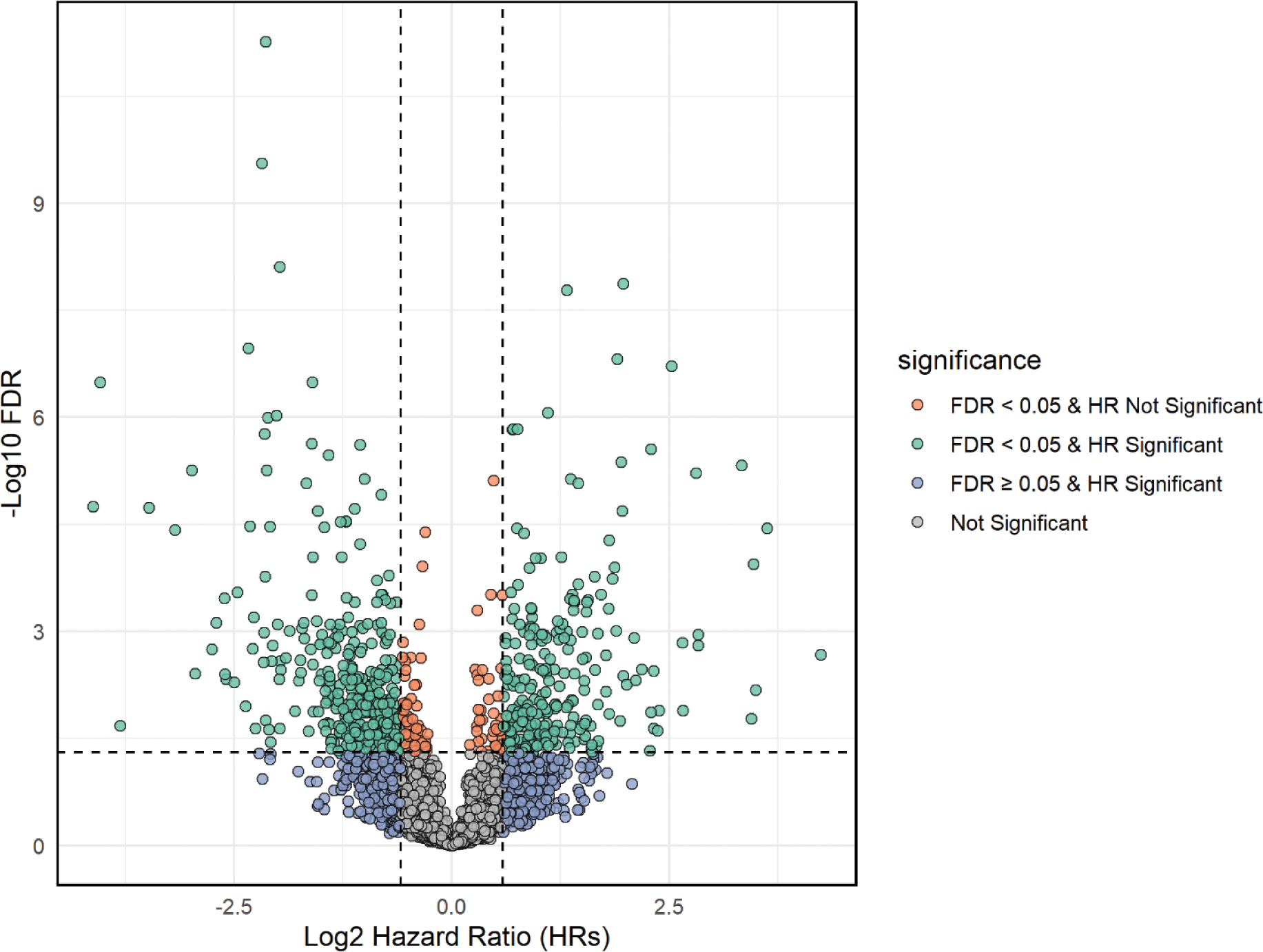
Results: Among 321 SLE patients recruited in the PRECISESADS project who were profiled at the transcriptional level, long-term data were available in 131 patients. Of those, 85 patients had a clinical SLEDAI-2K <6 and were considered for the present analysis. At the time of blood sampling, those 85 SLE patients had a mean (SD) age of 47.5 (13.9) years and a mean disease duration of 15.7 (9.7) years, and they were mostly women (n=84, 98.8%). The mean clinical SLEDAI-2K was 1.5 (1.5) and the mean total SLEDAI-2K was 3.6 (2.3), while 52 patients were in LLDAS (61.1%) and 26 (30.6%) in DORIS remission. At baseline, 59 (69.4%) patients were treated with hydroxychloroquine, 25 (29%) with synthetic immunosuppressants, and 36 (42.3%) with glucocorticoids at a mean daily dose of 2.3 (0.5) mg of a prednisone equivalent. After a mean observation time of 6.9 (2.6) years, flares had occurred in 30 patients (35%). Among those 30 patients, the first flare was developed after a mean time of 3.0 (2.0) years, and the non-cumulative count of those flares per domain was 18 articular, 9 cutaneous, 5 constitutional, 4 haematological, 3 vascular, and 2 renal flares. LLDAS was protective against flaring (Figure 1); no other baseline clinical variable was found to be predictive of flares. After correction for LLDAS, a total of 675 genes were associated with flaring with an FDR-corrected p <0.05 and an HR >1.5 (or an HR <0.667 for protective genes). Of those genes, 253 increased and 522 decreased the hazard for flare (Figure 2). Gene ontology analysis of positively associated genes showed that the GO:0045088 pathway (regulation of innate immune response) comprising the ZBP1 , PARP9 , TRIM21 , TRIM5 , LGALS9 , TRIM56 , HLA-B , ZNFX1 , TRAFD1 , ADAR , PARP14 , STAT2 , CLEC12B , CLEC7A , XIAP , and PAK1 genes was significantly enriched (FDR-corrected p=0.0018).
Conclusion: In our cohort of quiescent SLE patients, we corroborated that LLDAS is protective against flaring and demonstrated that biology is independently linked to disease recrudescence. In concrete, we found increased gene expression related to interferon stimulation ( STAT2 , TRIM21/Ro52 , TRIM5 , TRIM56 , ZNFX1 ), pyroptosis ( ZBP1 ), regulation of macrophage inflammatory activation ( CLEC7A , PAK1 , PARP9 , PARP14 ), T-cell activation ( LGALS9 ), increased RNA editing and autoantigen exposure ( ADAR ), and negative regulators of innate immune responses ( TRAFD1 ) to be associated with time to flare.
REFERENCES: NIL.
Acknowledgements: This work was supported by EU/EFPIA/Innovative Medicines Initiative (IMI) Joint Undertaking (JU) PRECISESADS grant no. 115565 and IMI 2 JU (now HIH) 3TR grant no. 831434.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (