

Background: Familial Mediterranean Fever (FMF) is the most common monogenic autoinflammatory disorder, characterized by recurrent fever and serosal inflammation, caused by mutations in the MEFV gene. This results in the unregulated activation of the pyrin inflammasome in myeloid cells. Colchicine is effective in preventing attacks, but in a minority of patients it can be insufficient for satisfactory disease control, and these colchicine-refractory individuals experience continuous hyperinflammation. Despite extensive research, our understanding of the specific phenotype of circulating immune cells in FMF patients remains limited, particularly regarding the molecular distinctions between colchicine-responsive and colchicine-resistant individuals. Single-cell RNA sequencing, alongside other high-resolution molecular techniques, would enables us to address these concerns, and to develop more targeted treatment strategies.
Objectives: This study aims to identify specific signatures of peripheral blood mononuclear cells (PBMCs) in colchicine-responsive and refractory FMF patients compared to healthy controls.
Methods: Peripheral blood samples were collected from FMF patients and healthy controls at Istanbul University. PBMCs were isolated and cryopreserved. For single-cell RNA sequencing, cells were thawed ensuring >80% viability, they were fixated, and the libraries were prepared using the Parse Bioscience Evercode Midi WT v2 kit. Libraries were quality-controlled with Agilent Bioanalyzer and sequenced on Illumina Novaseq 6000 S2 200 cycle flow cell, targeting 20,000 reads per cell. Raw reads were demultiplexed and aligned to the human reference genome (hg38) using the Parse Biosciences pipeline, spipe. High-quality cells were retained by filtering those with <250 detected genes, >20% mitochondrial expression, or genes expressed in <5 cells. Doublets were removed with DoubletFinder, and data were log normalized. Post-QC, 62,000 cells remained with a median of 2,500 genes per cell. Batch effects were corrected using Harmony. Clustering employed 35 PCs with a resolution of 0.6, and UMAP was used for dimensionality reduction. Immune populations were annotated via marker genes, and cluster counts generated pseudobulk profiles. Differential expression analysis (DESeq2) compared controls, colchicine-responsive FMF, and colchicine-refractory FMF.
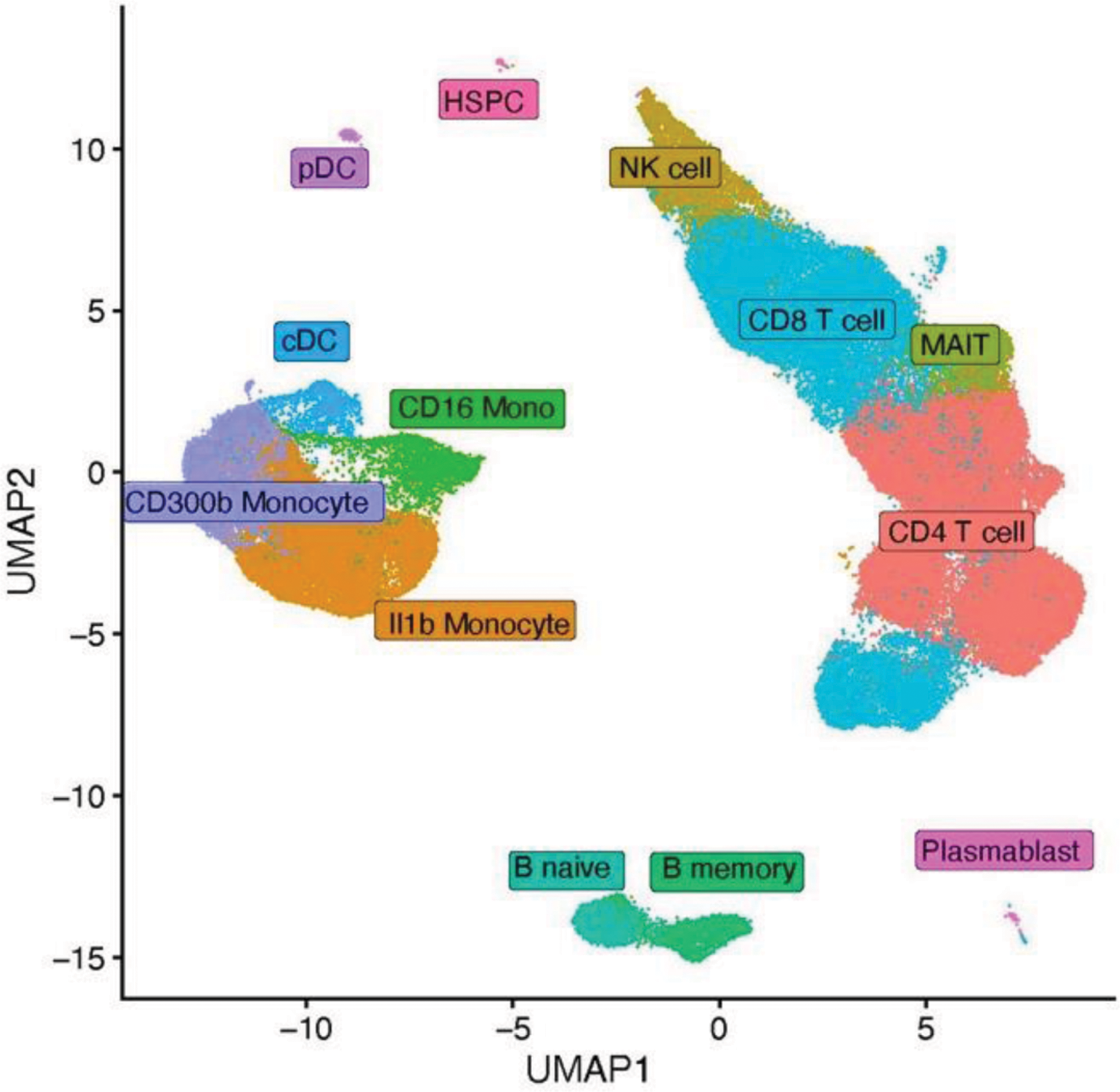
Results: The cohort included 15 participants: 10 FMF patients (6 colchicine-responsive, 4 colchicine-refractory) and 5 healthy controls. MEFV mutations in patients included M694V compound heterozygous and homozygous variants. Responsive patients received colchicine daily, while refractory patients were treated with canakinumab. Single-cell RNA sequencing identified 13 distinct PBMC clusters, corresponding to specific cell types based on marker expression (Figure 1). Most differentially expressed genes were observed in classical monocytes (CD14+ CD300+), activated classical monocytes (CD14+ IL-1β+), non-classical monocytes (CD16+), CD4+ T-cells, and CD8+ T-cells. In PBMCs, monocytes are the main pyrin inflammasome-expressing cells and likely harbour relevant differences in FMF. Classical monocytes of colchicine-responsive patients upregulate genes linked to chemotaxis and positive regulation of cytokine production pathways, while colchicine-refractory patients expressed more genes related to cytoplasmic translation and chemotaxis. Notably, activated classical monocytes of refractory patients express less leukocyte cell-cell adhesion, LPS response, and canonical NF-κB signal transduction related genes, alongside with increased cytoplasmic translation and ATP metabolic processes - features absent in responsive patients. Finally, non-classical monocytes showed downregulated immune response activating pathways in both patient groups. Colchicine-refractory patients upregulated cytoplasmic translation genes, while colchicine-responsive patients showed increased small GTPase and Ras signal transduction. These findings confirm inflammatory dysregulation in FMF monocytes, with some distinction between colchicine responsive and refractory patients, offering new pathways for future research. Lymphocytes also revealed significant differences. CD4+ and CD8+ T-cells in FMF patients upregulated oxidative phosphorylation and aerobic respiration related genes, compared to controls. Additionally, CD8+ T-cells showed upregulation of genes linked to cell killing capacity and NK-cell mediated cytotoxicity. These results indicate a potential metabolic shift that could contribute to disease progression.
Conclusion: This study extends previous findings of altered inflammatory gene expression in monocytes of FMF patients. Colchicine-refractory patients exhibited distinct monocyte profiles, while T-cell metabolic shifts were observed in all FMF patients. These findings lay the groundwork for further research into colchicine-refractory FMF, including epigenetic mapping and metabolic profiling, to develop targeted therapies.
UMAP plot of single cell RNA sequencing data obtained from PBMCs of FMF patients, resistant and refractory to colchicine and healthy controls. It identifies 13 clusters.
REFERENCES: NIL.
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (