

Background: Hypermobility Spectrum Disorders (HSD) are clinical conditions characterized by joint hypermobility [1] that exceeds normal ranges [2], accompanied by musculoskeletal and systemic symptoms that affect patients’ quality of life [3]. Although their clinical impact is recognized, the molecular pathways underlying HSD remain largely unknown. It is suspected that abnormalities in the protein structure of the extracellular matrix may contribute to the pathogenesis [4]. Furthermore, implications of immunological and epigenetic factors have been proposed [5]. However, the lack of specific biomolecular markers makes diagnosis and the development of targeted therapies difficult. Therefore, there is an urgent need for integrated research employing multiomics approaches to elucidate the molecular mechanisms of HSDs.
Objectives: Identify altered molecular pathways in patients with Hypermobility Spectrum Disorders through gene expression analysis.
Methods: In silico analysis of gene expression datasets from HSD patients was conducted using the GEO platform. The inclusion criteria for the datasets were: at least three cases diagnosed with HSD according to the 2017 criteria for hypermobile Ehler-Danlos syndrome/hypermobility spectrum disorders or the Villefranche criteria, and a minimum of three healthy controls. The GSE218012 and GSE77753 datasets met these criteria, comprising a total of 25 patients with HSD and 46 healthy controls. Differentially expressed genes (DEGs) were determined using PyDESeq2 in Python, considering as DEGs those with an adjusted p value < 0.05 and a log2 Fold Change of ±1. Pathway analysis was performed using the KEGG pathways database, and functional enrichment results were visualized with ShinyGo v0.8.
Results: A total of 425 differentially expressed genes were identified, of which 210 were upregulated and 215 downregulated. The upregulated DEGs (Figure 1A) were associated with a functional enrichment in pathways related to the formation of neutrophil extracellular traps and the cytokine-receptor interaction. SLE and IL-17 signaling pathways were also enriched. In contrast, the downregulated DEGs (Figure 1B) were mainly found enriched in the extracellular matrix-receptor interaction pathway and the TGF-beta signaling pathway.
Functional enrichment analysis of differentially expressed genes. A) Signaling pathways enriched for upregulated DEGs. B) Signaling pathways enriched for downregulated DEGs. The size of the points indicates the number of genes involved, and the color represents statistical significance of the enrichment.
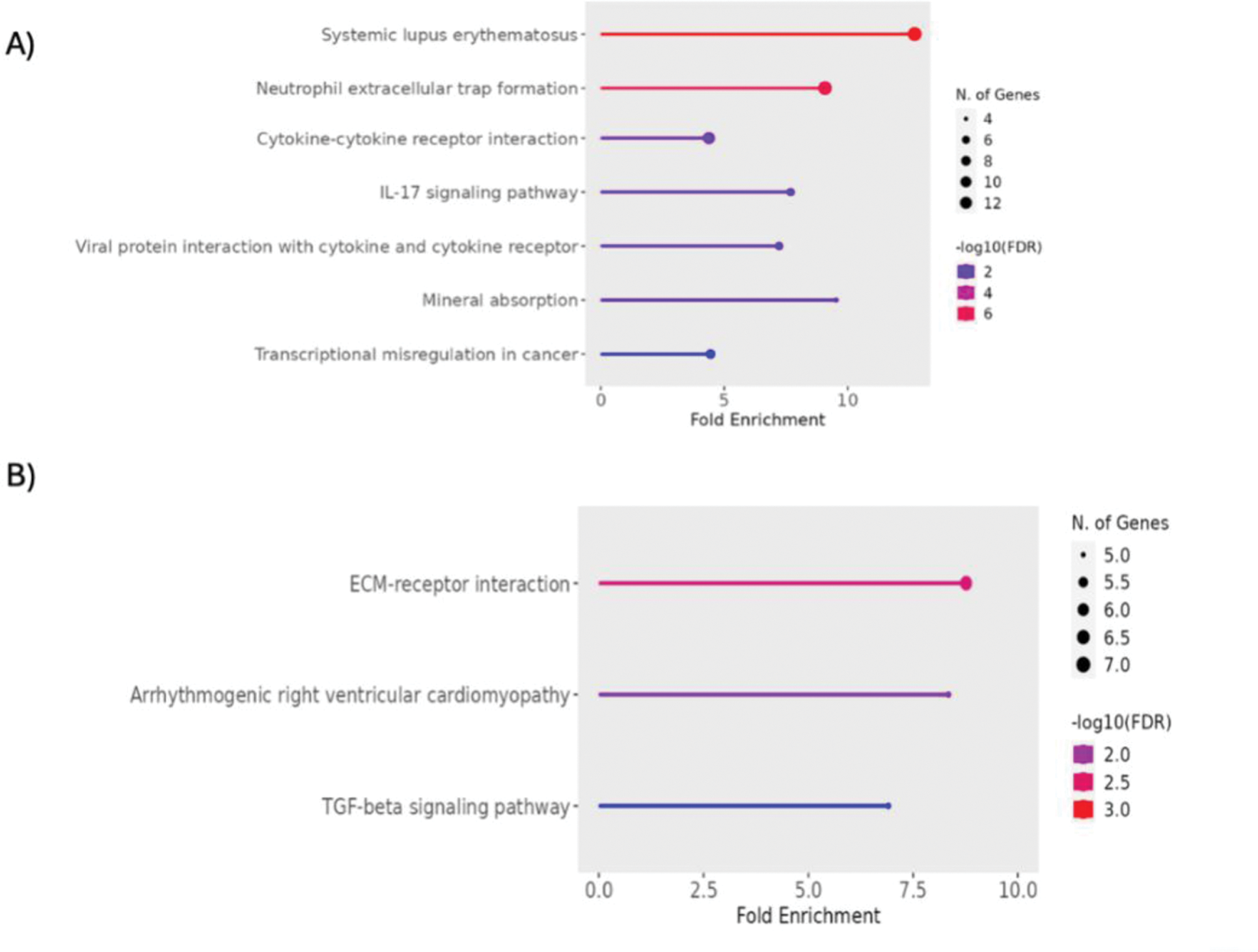
Conclusion: Gene expression analysis in patients with HSD revealed alterations in inflammatory and connective tissue remodeling pathways. The findings suggest that dysfunction in matrix-cell interaction and cytokine signaling contributes to the pathogenesis of HSD. These results provide valuable information for future research aimed at the identification of biomarkers and the development of targeted therapeutic strategies in patients with HSD.
REFERENCES: [1] Malfait F, Francomano CA, Byers PH, Belmont JW, Berglund B, Black JH, et al. The 2017 international classification of the Ehlers-Danlos syndromes. Am J Med Genet Part C-Semin Med Genet. 2017;175(1):8–26.
[2] Carroll MB. Hypermobility spectrum disorders: A review. Rheumatol Immunol Res. 2023;4(2):60–8.
[3] Morlino S, Dordoni C, Sperduti I, Clark CJ, Piedimonte C, Fontana A, et al. Italian validation of the functional difficulties questionnaire (FDQ‐9) and its correlation with major determinants of quality of life in adults with hypermobile Ehlers–Danlos syndrome/hypermobility spectrum disorder. Am J Med Genet. 2019;180(1):25–34.
[4] Chiarelli N, Zoppi N, Ritelli M, Venturini M, Capitanio D, Gelfi C, et al. Biological insights in the pathogenesis of hypermobile Ehlers-Danlos syndrome from proteome profiling of patients’ dermal myofibroblasts. Biochim Biophys Acta BBA - Mol Basis Dis. 2021;1867(4):166051.
[5] Gensemer C, Beck T, Guo L, Petrucci T, Morningstar J, Kornblau I, et al. Variants in the Kallikrein Gene Family and Hypermobile Ehlers-Danlos Syndrome [Internet]. 2024
Acknowledgements: NIL.
Disclosure of Interests: None declared.
© The Authors 2025. This abstract is an open access article published in Annals of Rheumatic Diseases under the CC BY-NC-ND license (